Follow our 3-Step Guided Tutorial to set up a RNA-seq Processed Counts experiment.
This is an interactive module built to prepare your first RNA-seq experiment from Processed Counts at your own pace.
To follow along with your own data, have your Processed Counts prepared to use for Part 1.
Download the demo files to follow along with the guided module and create an
RNA-seq experiment ready for exploration in the ROSALIND platform.
RNA-seq from Processed Counts
Step 1: Create a new Project folder by clicking ![]() . Enter a Title and Description (both can be edited at a later time).
. Enter a Title and Description (both can be edited at a later time).
Step 2: Select "Design a New Experiment" to access NanoString assay workflows, then click Next.
Step 3: Enter an "Experiment Title" and "Description" (both can be edited at a later time)
Then click Next.
Step 4: For Processed Counts files use the following selections:
"Method (Technology)" drop down menu to select "Sequence."
"Experiment Type" drop down menu, select "RNA-seq (Processed Counts)"
Then click Next .
Step 5: Use the toggle to select the Processed Counts file type, select "Raw"
Click the "Browse or Drop File" upload box to navigate to and select the demo Processed Counts file for upload, or drag and drop into the upload box to begin upload*. Once the file has successfully uploaded, a green checkmark will appear next to the file.

Click Next .
*If analyzing your own data, select your own Normalized Counts file for upload
Step 1: Make the following selections from the dropdown menus*:
Species: "Homo sapiens (human)"
Genome Build: "GRCh38"
*If analyzing your own data, select your own genome used
ROSALIND offers genome builds from NCBI and Ensembl. Unless comparing the experiment to historic data, it is recommended to select the most recent build from your preferred source (NCBI or Ensembl).
Click Next.
Step 2: Select the following from the dropdown menus*:
Kit Vendor: "NEB"
Kit Model: "NEBNext Ultra II RNA Library Prep Kit for Illumina"
*If analyzing your own data, select your own kit used
If you are unsure which kit was used for the experiment, contact your service provider to obtain that information. If the name of the kit is not listed, reach out to ROSALIND Support at support@rosalind.bio for guidance, or select "Other" to run a generic, unstranded analysis.
Click ![]() to confirm the selection, and ensure both a "Sample Kit" and "Library Kit" are registered to continue. An "Auxiliary Kit" is optional.
to confirm the selection, and ensure both a "Sample Kit" and "Library Kit" are registered to continue. An "Auxiliary Kit" is optional.

Step 3: From the drop down menu, enter that the Experiment contains samples that are biological replicates ("Yes" or "No"). Enter the "Total number of samples" in the Experiment: 24.
Click Next.
Step 1: Specify sample attributes by typing the attribute categories into the "Enter Attributes" line.
Enter the following attributes: "Treatment", "Time Course", and "Tissue"
Learn:
To learn how to create your own attribute file, download the blank experiment attribute file (.CSV format) by clicking ![]() .
.
Within the attribute csv file, attribute values can be manually assigned to each sample.
*If uploading your own data, create an attribute file for your experiment and upload in the next step.
Step 2: Upload the demo attribute file* into ROSALIND by clicking ![]() .
.
Click Next.
Step 3: ROSALIND provides a sample sheet to review for accuracy of sample information, changes can be made directly on the sample sheet if needed.
Once reviewed, click ![]() .
.
Learn:
*This option is only available for RNA-seq from FASTQ
Learn how to assign replicate groups. This is optional though recommended to visually enhance attribute tracking. Replicate group membership can also be used for attribute changes and tracking. If an error is noted and an edit is made, this edit will be applied to all samples in the same replicate group, reducing time for edits and risk of error.
Learn:
Learn how to create comparisons and covariate correction.
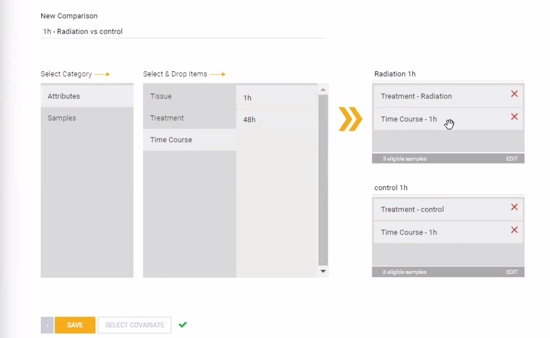
Comparisons can be configured now, prior to launching the analysis, or after the experiment has been processed and QC has been reviewed. To create a new comparison now, click ![]() . Comparisons can be created at the replicate group level (using the attributes) or individual sample level (using the sample names) by dragging and dropping different attributes or samples into the condition and control boxes on the right side of the page. Combinations of two or more different attributes can be used to create comparisons with specific subsets of samples. In the example image below, a comparison is set up across two treatment groups (Radiation vs Control) at a specific time point (1 hour). Alternatively, the comparison could be set up specifically at only the 48 hour time point, or including both time points simultaneously.
. Comparisons can be created at the replicate group level (using the attributes) or individual sample level (using the sample names) by dragging and dropping different attributes or samples into the condition and control boxes on the right side of the page. Combinations of two or more different attributes can be used to create comparisons with specific subsets of samples. In the example image below, a comparison is set up across two treatment groups (Radiation vs Control) at a specific time point (1 hour). Alternatively, the comparison could be set up specifically at only the 48 hour time point, or including both time points simultaneously.
Additionally, covariate correction can be added to the statistical model using the "SELECT COVARIATE" button. See this article to learn more about covariate correction methods.
Click the ![]() button to save the created comparison.
button to save the created comparison.

Demo Challenge:
Try creating the below comparisons with the demo data (or if using your own data, try creating a variety of comparisons as your study allows):
1) Condition vs Control:
D2 Antagonist vs Control
2) Condition vs Control with two attributes each:
48h - D2 Antagonist vs Control
3) Condition vs Control with Covariate Correction (2 comparisons will be created if set up correctly):
Radition vs Control - Time Course
C Radition vs Control - Time Course
*Note: If comparisons are set up correctly, ROSALIND Intelligence will automatically name the comparison as shown in the image.
Once all comparisons have been saved, click Next.
Learn:
Learn how to upload FASTQ files. For more guidance on FASTQ file upload, see our Formatting Guides.
Do not close or navigate away from the page until import is complete.

After successful import, the experiment will automatically launch. ROSALIND will notify you via email once the processing is complete and your experiment is ready for exploration.

RNA-seq from Processed Counts
Step 1: Navigate to and select the project folder containing the created "RNA-seq Processed Counts Demo" experiment*. Then click into the experiment.
*If your own data was used, click into your applicable experiment
Step 2: Explore the experiment summary tab (page icon, default)![]() for the experiment.
for the experiment.
*Note: RNA-seq data from Processed Counts will not generate a QC tab, a Genomic Region Notes plot, or a Gene Body Bias Notes plot.

Identify the location of the
1) Experiment title
2) Experiment description
3) Experiment setup parameters
4) Methods and citations

Step 3: Click on the Sample Correlation heatmap to open the figure in a larger window.
Evaluate the correlation of your sample correlations in relationship to defined attributes.
Use the ![]() and
and ![]() options to further explore the plot options.
options to further explore the plot options.
Use these features to further explore each plot.

Step 4: Click on the Violin plot. Select the "Additional Images & Videos" section on the bottom left to learn more about how to interpret this plot.

Step 5: Click on the MDS plot and determine sample similarity between defined attributes.
Step 1: Select the Samples tab at the top (test tube icon) ![]() to navigate to the Samples Page.
to navigate to the Samples Page.
Step 2: Explore the Samples page.
Demo Challenge:
Locate the following options on your page:
1) Verify all attribute information is recorded in the table
2) ![]() to add additional attributes for differential analysis post processing
to add additional attributes for differential analysis post processing
*Note: This option is only available to the owner of the experiment
Step 1: Select the File Management tab at the top (Cloud icon) ![]() to navigate to the File Management Page.
to navigate to the File Management Page.
Step 2: Explore the different file options for download. Examples include:
1) Source Files Counts Matrix (RNA-seq Processed Counts file)
2) Attribute file
3) Comparison Results (Differential expression results files)
RNA-seq from Processed Counts
Step 1: Navigate to and select the project folder containing the "RNA-seq Processed Counts Demo" experiment*. Then click into the experiment.
*If your own data was used, click into your applicable experiment
Step 2: Select the Discovery and Analysis Tab ![]() at the top.
at the top.
Step 3: Explore the Discovery and Analysis page
Locate the following items on your page:
1) How to create new comparisons with ![]()
2) How to create new meta-analysis with ![]()
3) Normalized Expression
4) Term Exploration

Step 1: Click into the Normalized Expression Dataset

Step 2: Explore how to visualize data for single targets, multiple targets, or a gene list in multiple views
*Note: To select multiple targets, hold CTRL or CMD and click targets of interest

Step 1: Click into Term Exploration to explore Gene Set Variation Analysis (GSVA) Results

Step 2: Explore how to visualize GSVA pathways and scores as a box plot, bar graph, heatmap, or data table.

Step 1: Click into the "48h - D2 Antagonist vs Control" created differential analysis comparison

Step 2: Identify the significantly differentially expressed genes on the left-hand side and the default parameters determined by ROSALIND Intelligence.

Step 3: Explore the volcano plot interactivity.

Step 4: Toggle the sample normalized expression chart between individual samples and sample groups box plot.

Step 5: Explore the heatmap interactivity.
Custom Visualizations: To create a custom volcano plot and heat map, hold down the control key on a PC keyboard or command key on a MAC keyboard, while clicking multiple genes from the list on the left or select a pre-created gene list (see this article for more information on how to create a gene list).
Custom Tabs: The tabs on the left margin of the significant gene list allow for visualization and exploration customization. Explore each tab:
Filter tab ![]() : represented by the funnel icon, allows the option to switch between created
: represented by the funnel icon, allows the option to switch between created

filters or create new filters. Select the plus "![]() " icon to create a new filter.
" icon to create a new filter.
Within the Create a New Filter page, enter a new filter name, cut-off values, adjusted or unadjusted p-value option and optional color icon. Select ![]() to update the table on the right with applicable genes according to the filter parameters.
to update the table on the right with applicable genes according to the filter parameters.
Continue adjusting the parameters until satisfied with the gene output, then click ![]() .
.
The new filter and associated interactive analysis output will begin processing. You will receive an email to begin exploring the data when processing is complete.
Group tab ![]() : represented by three horizontal lines, allows grouping of significant genes.
: represented by three horizontal lines, allows grouping of significant genes.
Select whether to group genes by "Clusters" or "Not Grouped".

Then select how to sort the gene list.

Search tab ![]() : represented by a magnifying glass, opens a search bar that can be used for a gene or list of genes.
: represented by a magnifying glass, opens a search bar that can be used for a gene or list of genes.
Color Pallet tab ![]() : represented by a colored circle, allows customization of the heatmap color scheme.
: represented by a colored circle, allows customization of the heatmap color scheme.
Find your favorite heatmap color.
Custom Pathways: Explore the pathway table on the right.
ROSALIND Intelligence Enrichment Summary: lists the key pathways and terms that are the most representative of the important biological changes in your system from across all 50 knowledge bases.
*Note: if less than 50 significant pathways are found within your comparison, the ROSALIND Intelligence algorithm will not run and the Summary will simply display the top 50 pathways ranked by adjusted p-value.

Pathways Summary: Specific pathways can be selected from the drop-down menu from across 50 knowledgebases.

Click a pathway of interest to explore. Note how all visualizations change upon selection of a pathway to facilitate deep exploration.

Knowledgebase Deep Dive: Pathways can be visually explored on a deeper level by clicking the magnifying glass ![]() on the pathways summary page. A complete view of results for each knowledgebase can be explored by clicking on the magnifying glass on the pathway summary page.
on the pathways summary page. A complete view of results for each knowledgebase can be explored by clicking on the magnifying glass on the pathway summary page.

Explore the full results for a knowledgebase by altering its Chart Type, Sort By, and Color menu options.

To view pathway diagrams and visualizations, select the Wikipathways knowledgebase under the "List Type" drop down menu. Then click on the Gold magnifying glass ![]() for a pathway of interest to view the corresponding diagram.
for a pathway of interest to view the corresponding diagram.
To return to the interactive analysis dashboard, select the differential analysis tab ![]() at the top of the page.
at the top of the page.

Volcano Plot/Box Plot: To download the volcano plot or box plot, click on the visualization to open up a new window with options for download.
Heatmap: To download the heatmap, click on the heatmap to initiate an immediate download to your hard drive.
Lists and select views: To download lists such as filter views and pathway visualizations, select the cloud icon ![]() to initiate download.
to initiate download.
*Note: Some larger images may take longer to download. Clicking quickly or on multiple images will initiate multiple downloads which may be blocked by your browser. For more information on how to adjust these settings, see the FAQ section of this article.