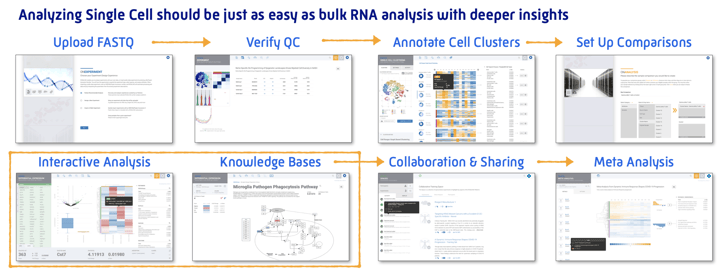
Reimagining single cell analysis from import to interpretation as an intuitive web experience
ROSALIND transforms the analysis of Single Cell RNA-Seq with an end-to-end web-based experience for analysis, interpretation and collaboration. Interactive analyses of single cell clusters reveals biology of cells.
Automated clustering provides Seurat methods, so you can choose the clustering that fits best
Assisted cell type identification saves time with integrated knowledge bases
Dynamic plots interactively display T-SNE, U-Map, Bar Charts, Donuts and Heatmaps to explore single cell trends
Native collaboration capabilities facilitate data sharing and teamwork
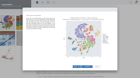
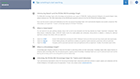
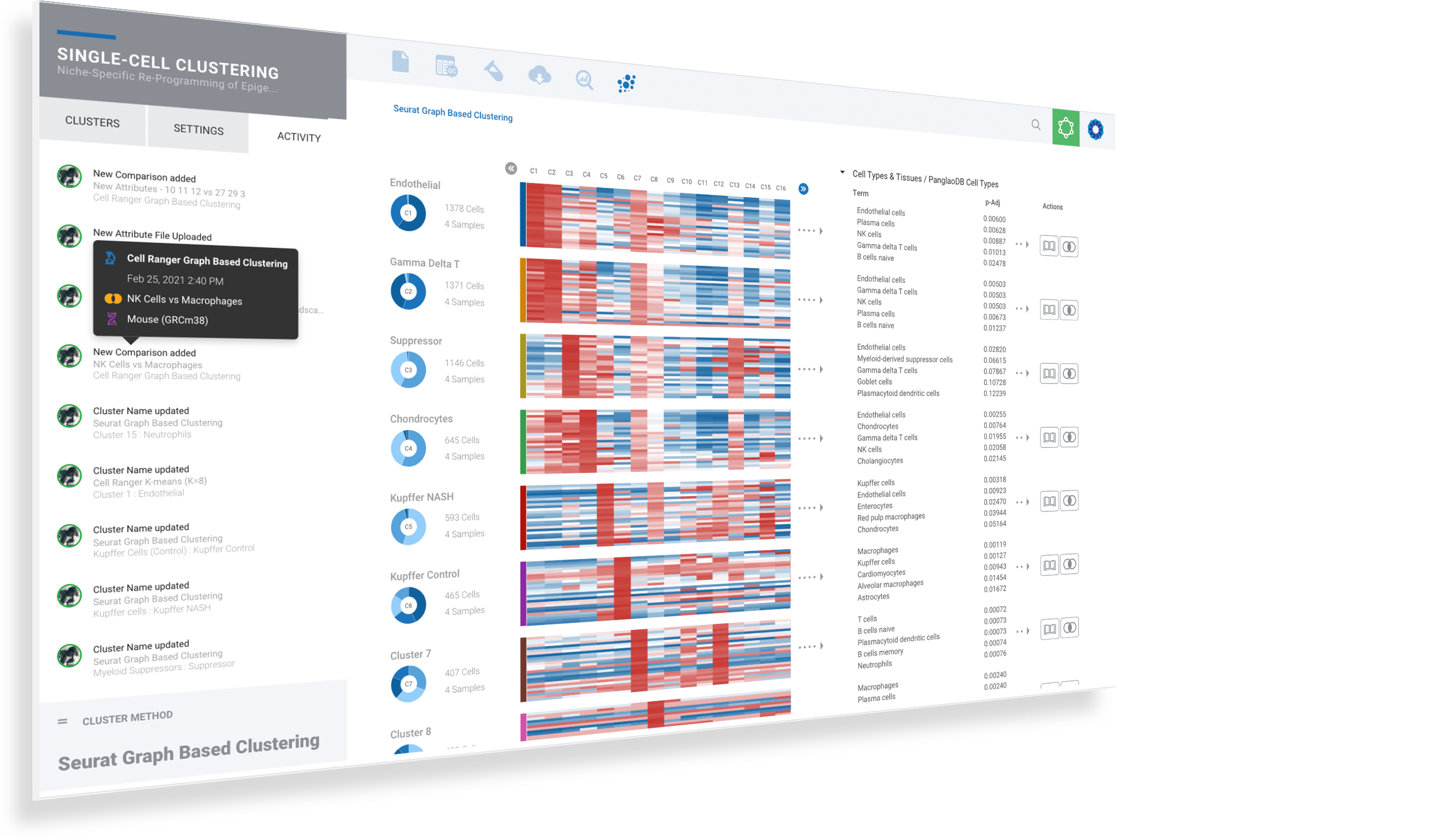
Annotate single cell data seamlessly & interactively
ROSALIND intelligently gathers relevant knowledge to help you best identify & annotate cell types. Notes are securely maintained across cluster methods, so you can capture observations in any context without losing a beat.
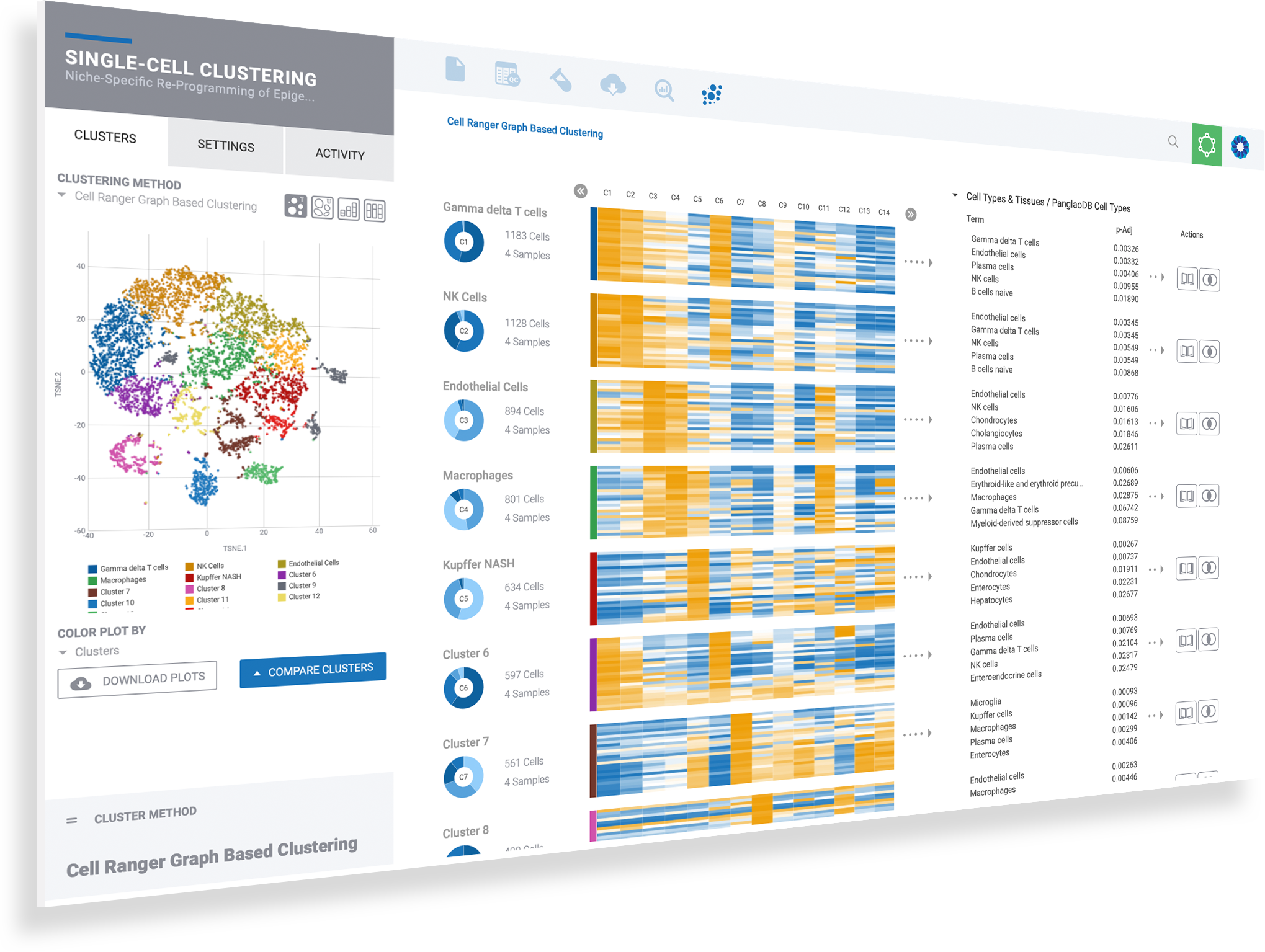
Select clusters to see heatmaps with marker genes
Toggle dynamic plots to see cells by sample, or cluster
Adjust settings to see 10, 25 or 50 marker genes, change colors or customize plots
Mouse over donut plots to see sample mix & cell counts
Click on Cell Types to explore knowledge base results for assisted identification
Use mouse gestures or scrolling on the T-SNE & UMAP to zoom and pan across the plot
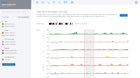
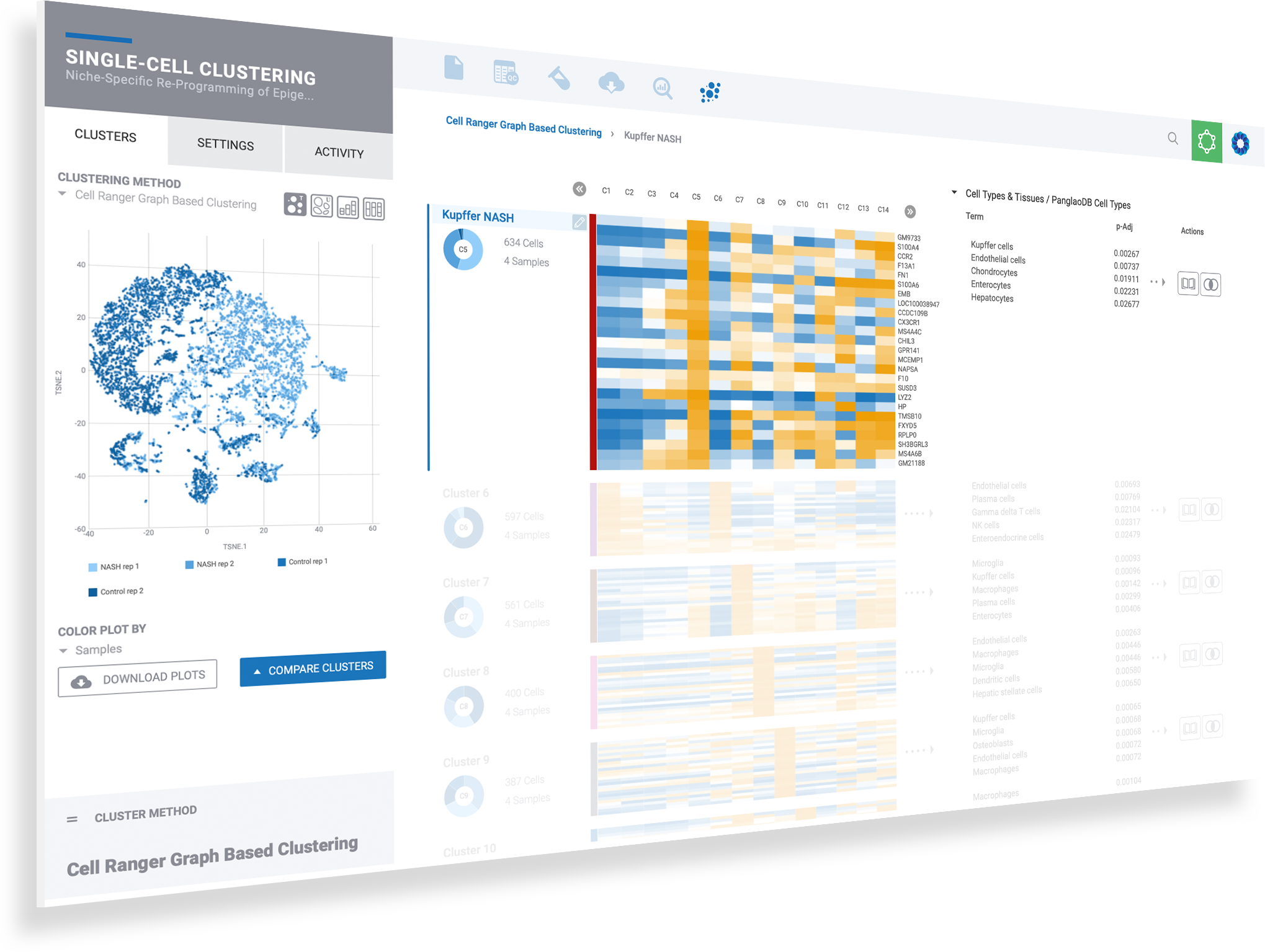
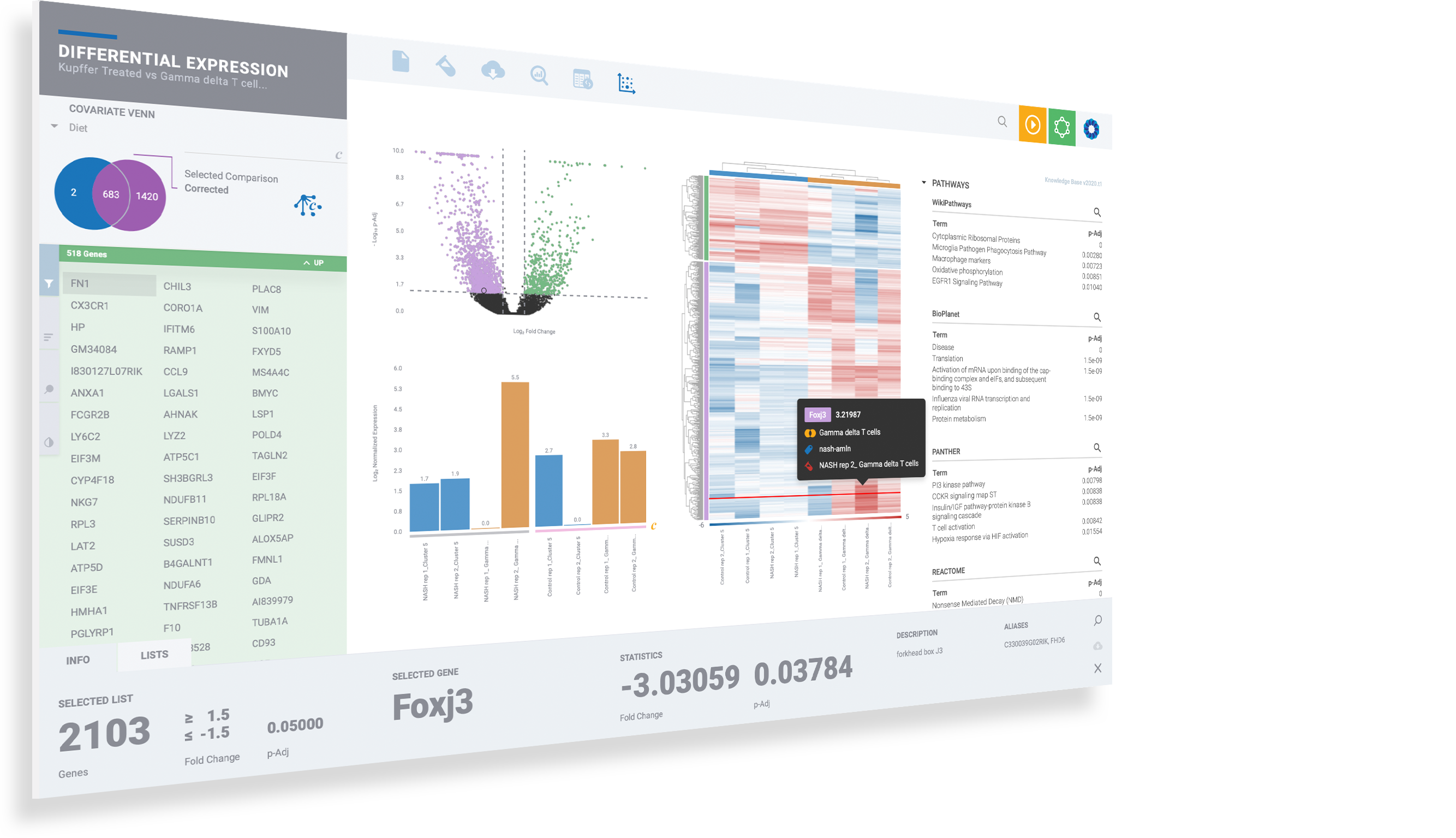
Visualize expression of marker genes & setup comparisons across any combination of clusters
Dive deeper into single cell clusters to explore gene expression using violin plots and interactive T-SNE and UMAP projections. Define cluster comparisons to see differentially expressed genes & pathway interpretation.
Immersive information on every data point helps to guide scientific inquiry
Select any gene to instantly visualize expression across every cluster
Create new comparisons by selecting desired cell clusters
Perform comparisons for any clustering method to see results from Seurat clustering, independently
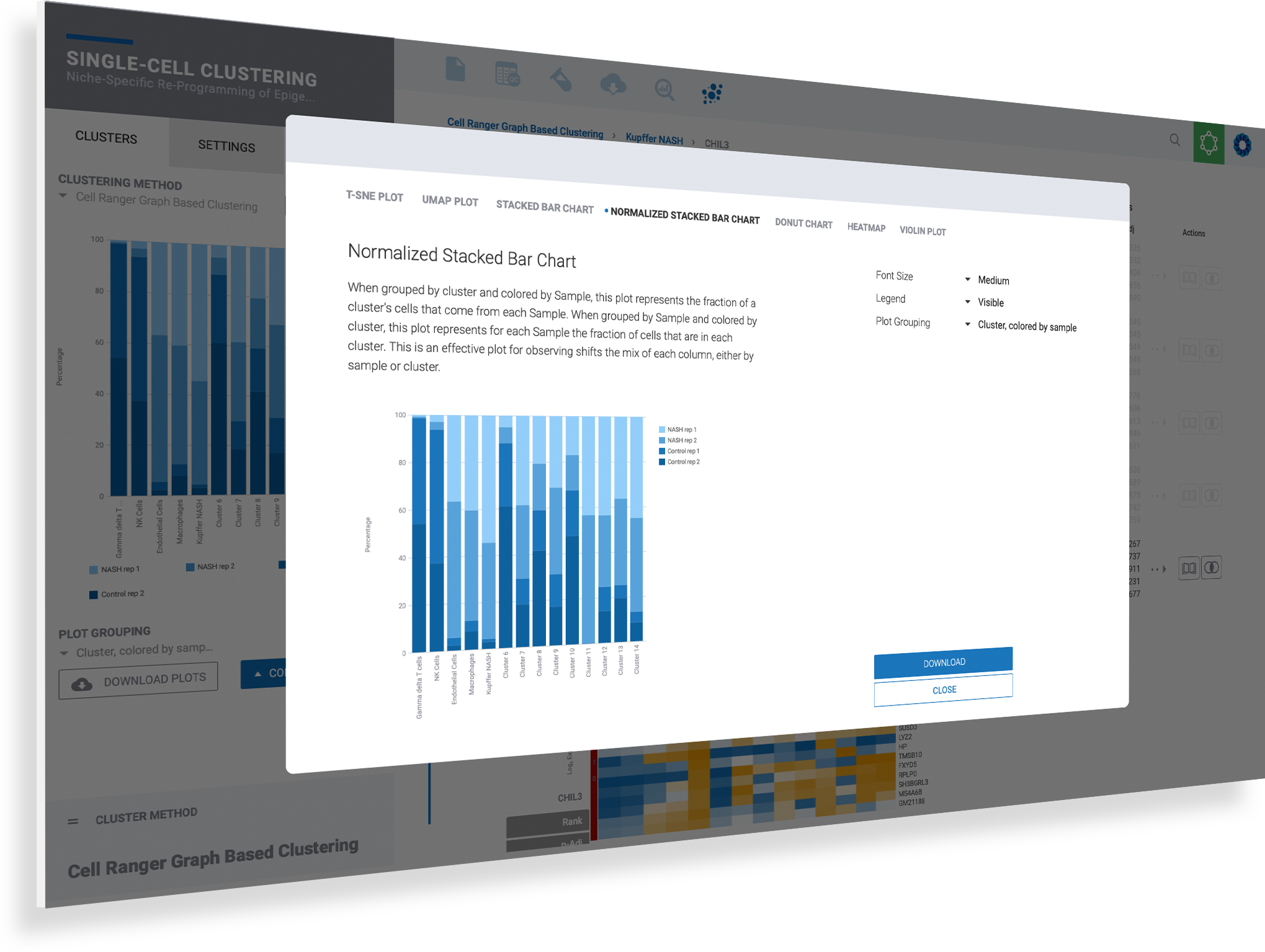
Customize and download any plot for presentation with publication-ready figures
Analyzing your data is only half the battle. Getting the right plot and image to communicate your findings makes all the difference. ROSALIND simplifies single cell data visualization giving you the plots you need with the controls to adjust fonts, plot sizes, zoom into clusters, hide legends, and see results across all samples.
Customize each plot based on the context of your data to see genes, clusters, or samples
Use the navigation menu in the enhanced plot control center to toggle plot types
Fine-tune your plots using the options provided uniquely for each plot type
Collaborate on single cell analysis & annotation
Single cell provides unprecedented data and granularity into biology and challenges our very understanding. Accelerate your single cell interpretation by collaborating in real-time with scientists across the globe.
Interactive activity feeds capture contributions and provide attribution to each team member
See changes to cluster annotation, sample annotation, cluster comparisons, gene cut-offs and more
Check out profile details for your collaborators and teammates
Quickly assess each collaborative contribution by mousing over the activity to see highlights
Instantly navigate to any activity by clicking on the item
Analyze single cell clusters just as easily as bulk RNA-Seq with the same powerful analysis and even deeper insights
Enjoy the thrill of seeing your differential expression results come to life as dynamic, interconnected plots and diagrams with gene lists, heatmaps, volcano & MA plots, and advanced covariate correction.
Expand the controls on the left to change color schemes, gene sorting and to create new filters and cut-offs
Use the bottom bar to pertinent gene information at your fingertips, including gene list management
Command-click or Control-Click (PC) to multi-select genes and customize plots
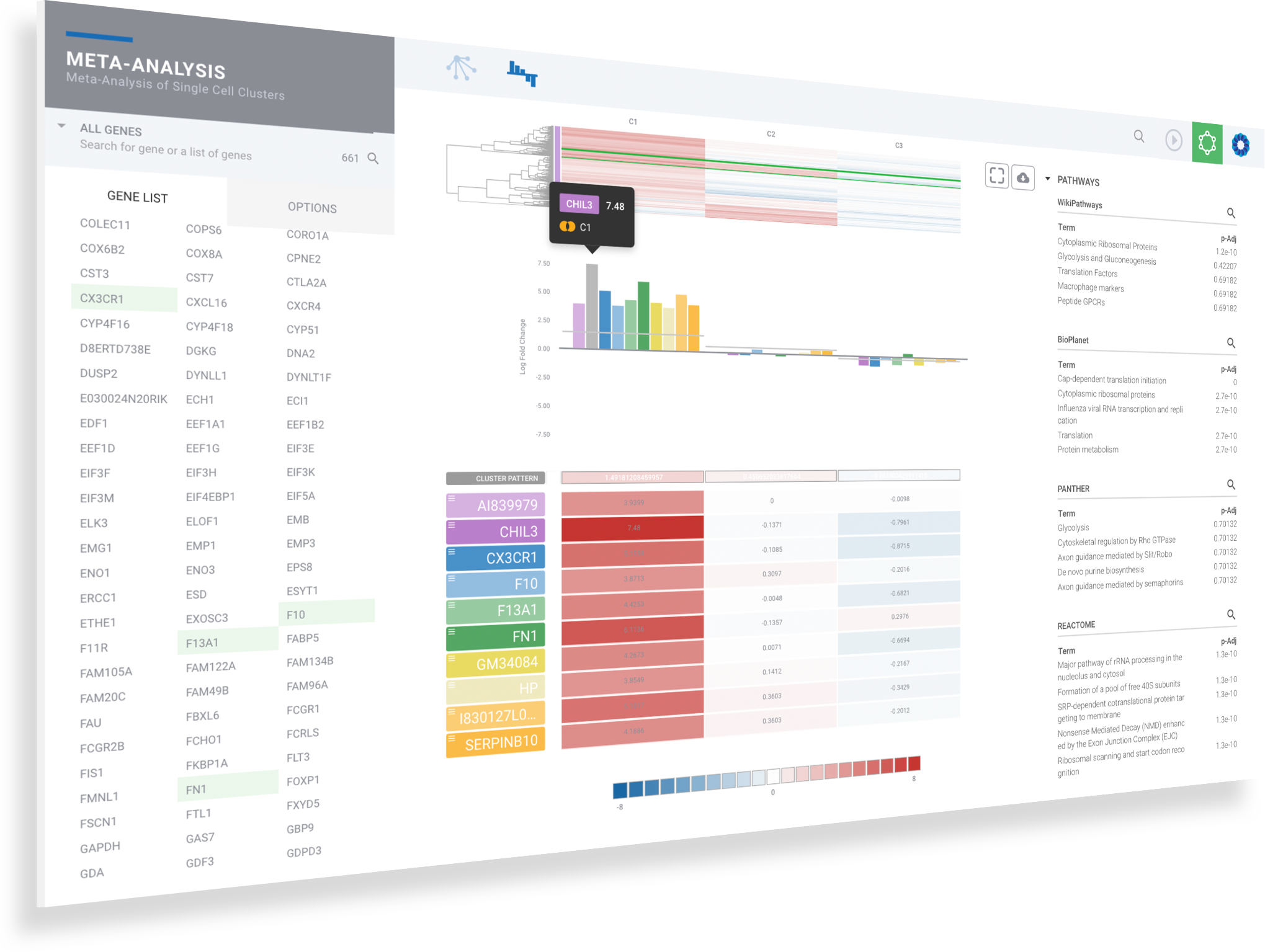
Evaluate top pathways and GO terms amongst 50+ knowledge bases by click on terms to instantly evaluate signature expression levels
Click on magnifiers to access additional details
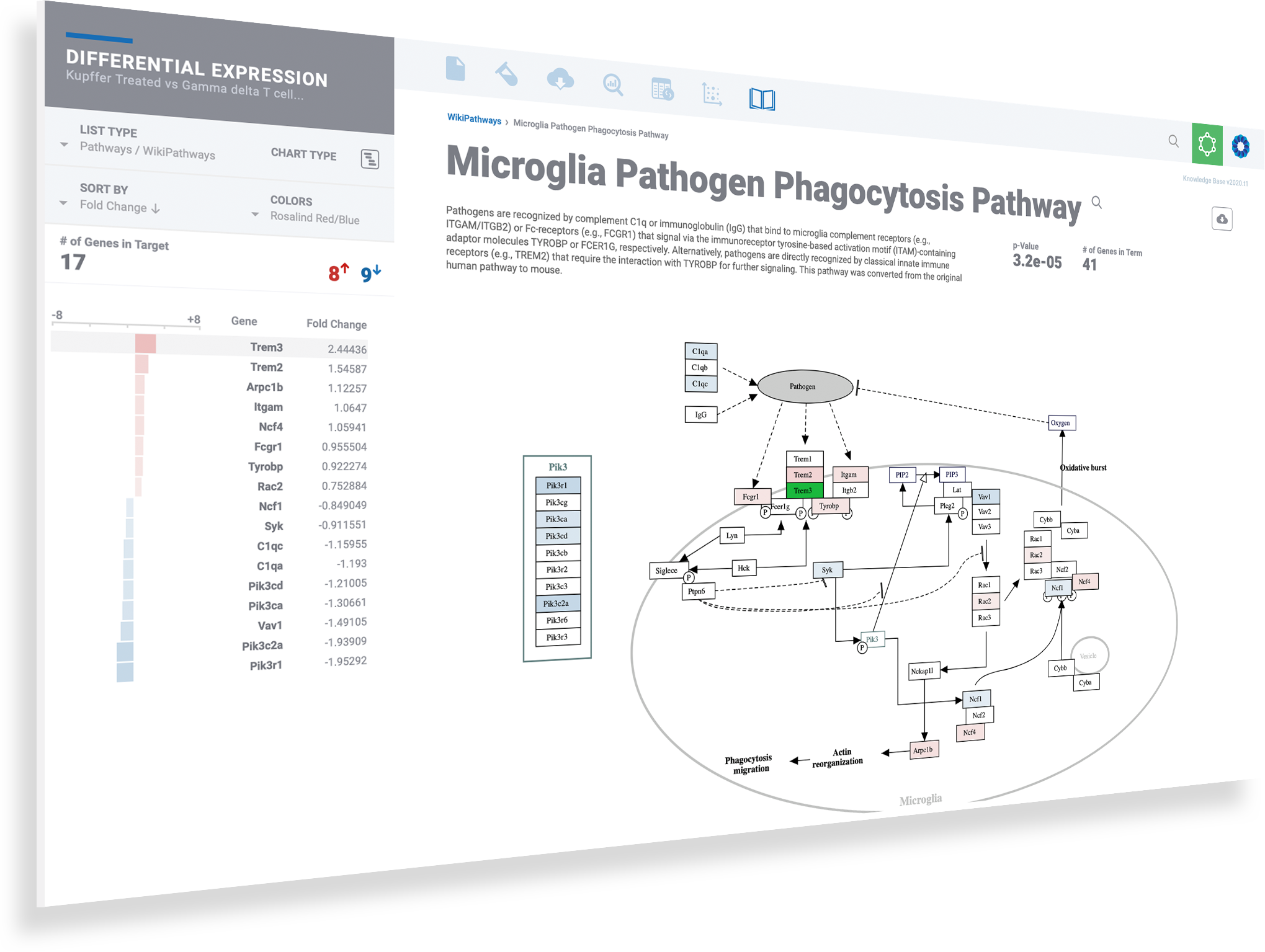
Dive deeper into pathways & the networks connecting them
Discover more and save valuable resources through the integrated pathway interpretation provided by the ROSALIND Knowledge Graph and integrated biomedical knowledge bases. Explore interactive pathway diagrams with detailed term descriptions, annotated fold-change colors, and gene heatmaps.
Explore interactive pathway diagrams to evaluate gene expression levels across pathways
Toggle knowledge bases with the list menu on the left
Click the download button to access publication-ready pathway diagrams as well as the associated heatmap and bar plots
Use breadcrumbs in the header to navigate back to prior levels for gene, term and knowledge base resources
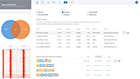
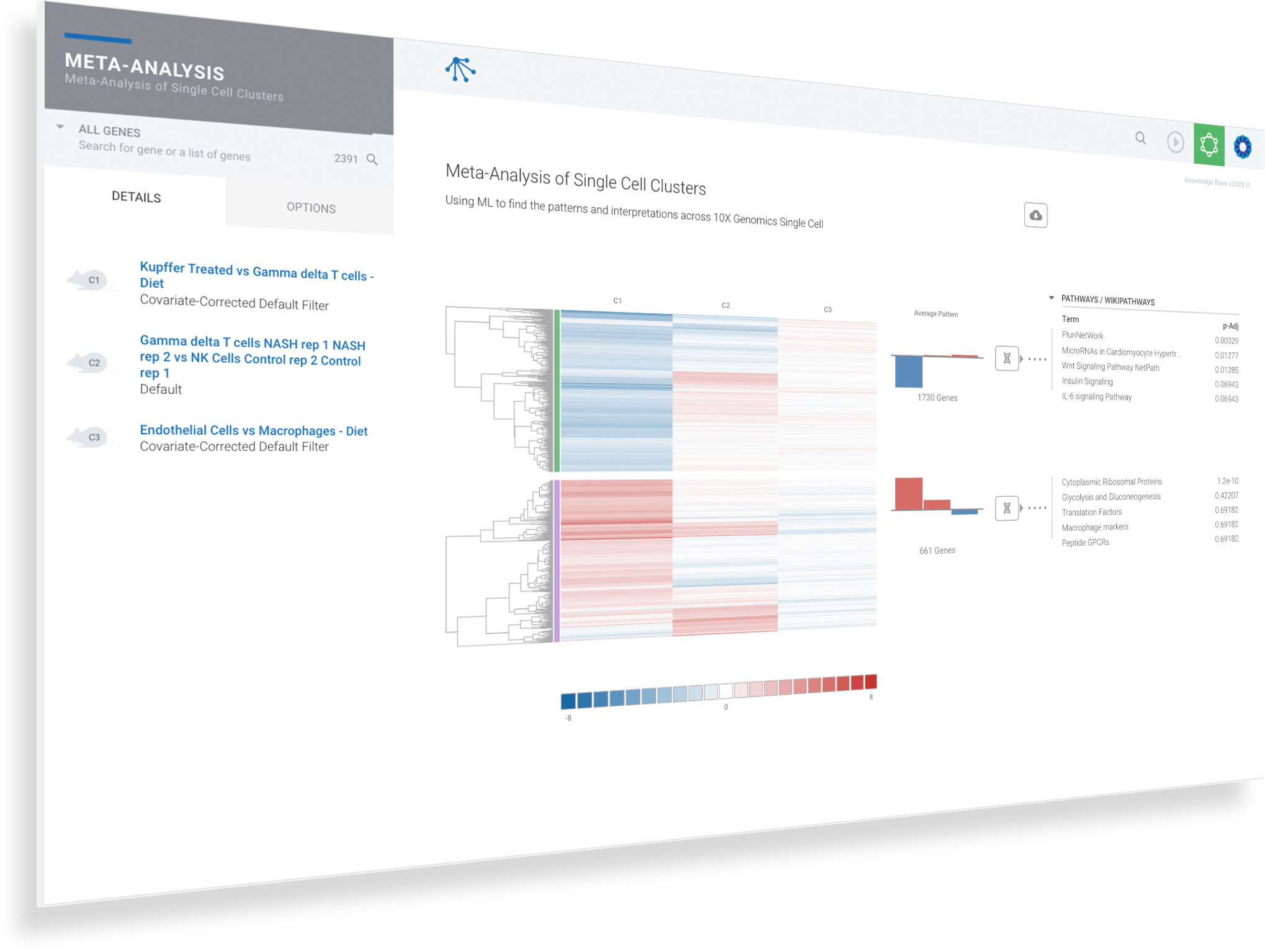
Harness machine learning to analyze single cell clusters across comparisons, experiments and multi-omics datasets
ROSALIND Meta-Analysis combines multi-omics datasets and uses unsupervised machine learning coupled with our advanced Knowledge Graph to intelligently uncover and interpret gene signatures
Single cell experiments are easily compared and contrasted with RNA-Seq, NanoString, ATAC-Seq and ChIP-Seq experiments
Up to 50 multi-omics datasets may be merged in a single meta-analysis
Navigate to and explore underlying experiment details by clicking on the comparison
Navigate to meta-analysis details to access advanced interpretation tools by clicking on the genome icon for the desired siganture





















/Rosalind_Logo_Primary_RGB.png)