How to Analyze Gene Expression with ROSALIND
Empowering Scientists to Accelerate Time to Discovery
WHY STUDY GENE EXPRESSION or RNA-SEQ?
The study of gene expression provides valuable insights into the nature of diseases and the effect of treatments by quantifying the activity of RNA in a biological sample. RNA-seq is a fast-growing Next Generation Sequencing (NGS) assay for evaluating gene expression, alternative splicing transcripts and fusions.
Scientists working in Oncology, Immunology, Regenerative Medicine, Drug Discovery and other areas of research often conduct experiments between healthy and disease states to identify Differentially expressed genes and biological pathways to discover therapeutic targets. Comparisons between these differential patterns reveal unique gene signatures valuable for drug and diagnostic development.
OVERVIEW
ROSALIND is a cloud platform that connects researchers to experiment design to quality control, differential expression and pathway exploration in a real-time collaborative environment.
Scientists of every skill level benefit from ROSALIND since no programming or bioinformatics are required. By accepting raw FASTQ sequence data as well as processed counts data, ROSALIND enables powerful downstream analysis and truly insightful visualizations on gene expression datasets. Receive same-day results with every experiment in an interactive experience designed for ease of use and saving valuable time.
HOW TO ANALYZE DIFFERENTIAL GENE EXPRESSION
ROSALIND enables scientists and researchers to analyze and interpret differential gene expression without the need for bioinformatics or programming skills. All that is required is basic background in biology and a current subscription or active trial.
Biological questions can also be explored independently, or in conjunction with, uploaded experiment data as ROSALIND automates the import of public data from the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) and Gene Expression Omnibus (GEO).
DISCOVERY CAPABILITIES

FOUR STEPS TO SUCCESS WITH RNA-SEQ
ROSALIND simplifies data analysis and works like a data hub interconnecting every stage of data interpretation. The ROSALIND Gene Expression discovery experience enables visual exploration and self-investigation of experiment results to give researchers the freedom to adjust cut-offs, add comparisons, apply covariate corrections, and even find patterns across multiple datasets, without the need for bioinformatic expertise. There are four easy steps to performing RNA-seq data analysis on ROSALIND.
1. EXPERIMENT DESIGN
Starting an RNA-seq data analysis begins with creating a new experiment and capturing the experiment design. ROSALIND walks through the key aspects of an experiment in a guided experience to record biological objectives, sample attributes and analysis parameters. These details become the basis of the experiment discovery dashboard. Researchers who publish papers and work with NCBI public data know the importance of natively supporting NCBI data models. ROSALIND fully supports the NCBI BioProject and BioSample models for metadata assignment and sample attribute descriptions. ROSALIND also enables scientists to create custom attributes to describe biological behaviors in terms relevant to the experiment. Setup of comparisons is simplified by describing and annotating samples using these familiar terms. This methodology minimizes the risk of differential expression errors when selecting samples for comparison.
For RNA-seq data analysis, ROSALIND provides scientists with a choice: a) Begin with raw FASTQ files produced by high throughput sequencing, or b) Use processed data files generated by another analysis pipeline. Processed data is imported as normalized or raw counts. This provides flexibility for scientists to utilize the ROSALIND discovery experience to visualize and interpret data regardless of the data source. When analyzing raw FASTQ files, ROSALIND streamlines data analysis using an advanced pipeline for analysis that includes intelligent quality control with automatic contamination detection, identification of Differentially expressed genes and deep pathway interpretation. Visit the technical specifications section to learn more about the ROSALIND RNA-seq data analysis pipeline and available reference materials.
For proper RNA-seq results, an analysis pipeline must adjust for sample preparation and proprietary differences in library preparation kits used in the experiment. Not only is the kit selection important for targeting and capturing the desired transcriptomic elements, the analysis pipeline adjusts and optimizes for the kit’s unique characteristics, such as strandedness, strand direction, any unique molecular identifiers (UMIs) as well as the adapters used. ROSALIND integrates and supports a broad library of sample and library preparation kits, automatically calibrating each analysis with the appropriate details. To learn more about supported kits, visit the technical specifications section. Featured kits and instrument partners are also listed below.
2. RNA-SEQ QUALITY CONTROL
Researchers must be confident in the quality control phase before gathering insights from an RNA-seq experiment, otherwise the results of the analysis should not be trusted. Biology’s mysteries are elusive and complex. Time should not be lost chasing corrective measures for outliers, contamination, swapped samples and the many other errors that can occur in the course of a well-designed experiment.
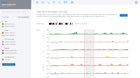
Some of the most important Quality Control metrics to verify are Q30 scores, alignment rates, ribosomal content, duplicate rates, sample correlation, gene coverage, genomic regions and multidimensional scaling (MDS) or principal component analysis (PCA) for all samples. When ROSALIND detects low alignment, non-aligning reads are evaluated for possible contamination. If ribosomal content is higher than expected, ROSALIND generates alerts. With Illumina sequencers, the results are usually good when Q30 values are over 85% and alignment rates are over 80% for the target species. Additionally, duplication rates less than 25% with fewer than 10% of reads trimmed is preferred. Researchers can eliminate offending samples and the deleterious effects on results by identifying the sample as an outlier and move confidently into the discovery and exploration phase of results interpretation.
ROSALIND Quality Control Intelligence identifies potential data quality issues and triages the data before presenting the results. This eliminates the needs for researchers to be experts in Sequencing quality control issues. Learn how researchers gain confidence in their results through Quality Control Intelligence.
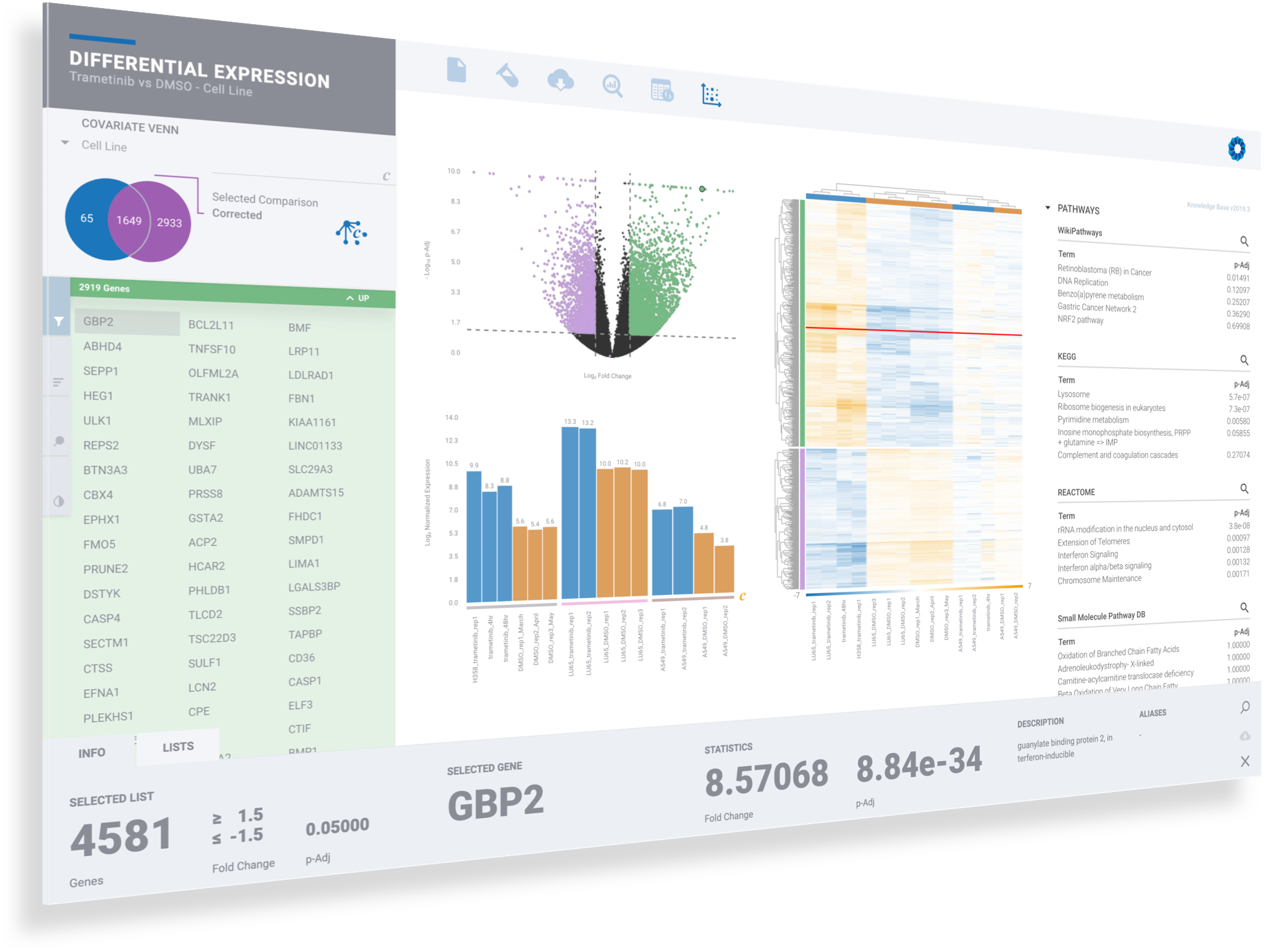
3. ANALYSIS & DISCOVERY
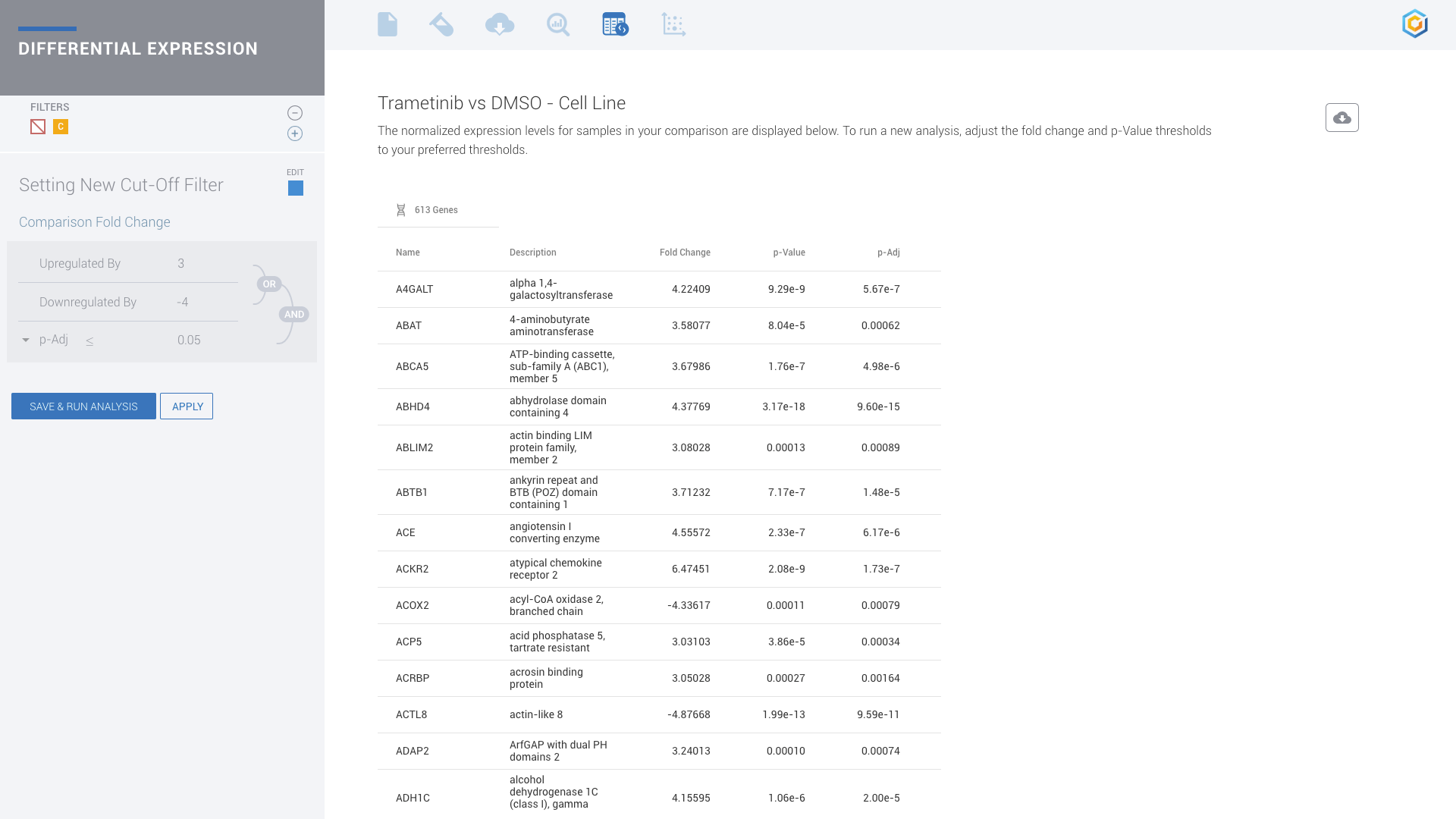
A typical RNA-seq analysis provides a list of Differentially expressed genes, generally in the form of a massive and obtuse CSV file. Unfortunately, this often results in more questions than answers for scientists. Multiple applications may also need to be used to generate this CSV file. Such applications often have a wide range of complexity with non-standard input/output formats, many of which are command-line tools requiring advanced knowledge in programming — an exercise well beyond the level of most biologists.
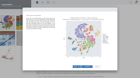
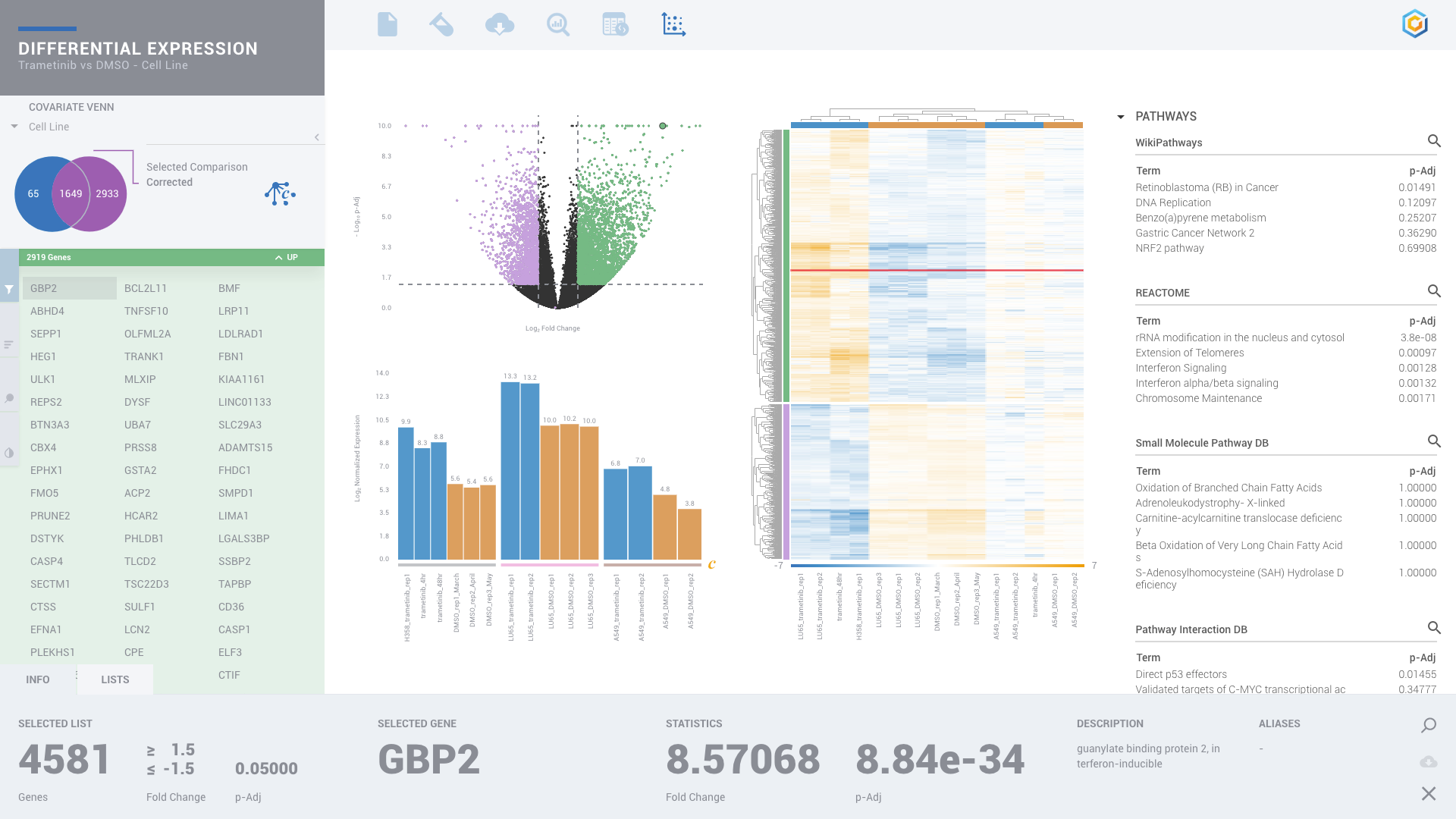
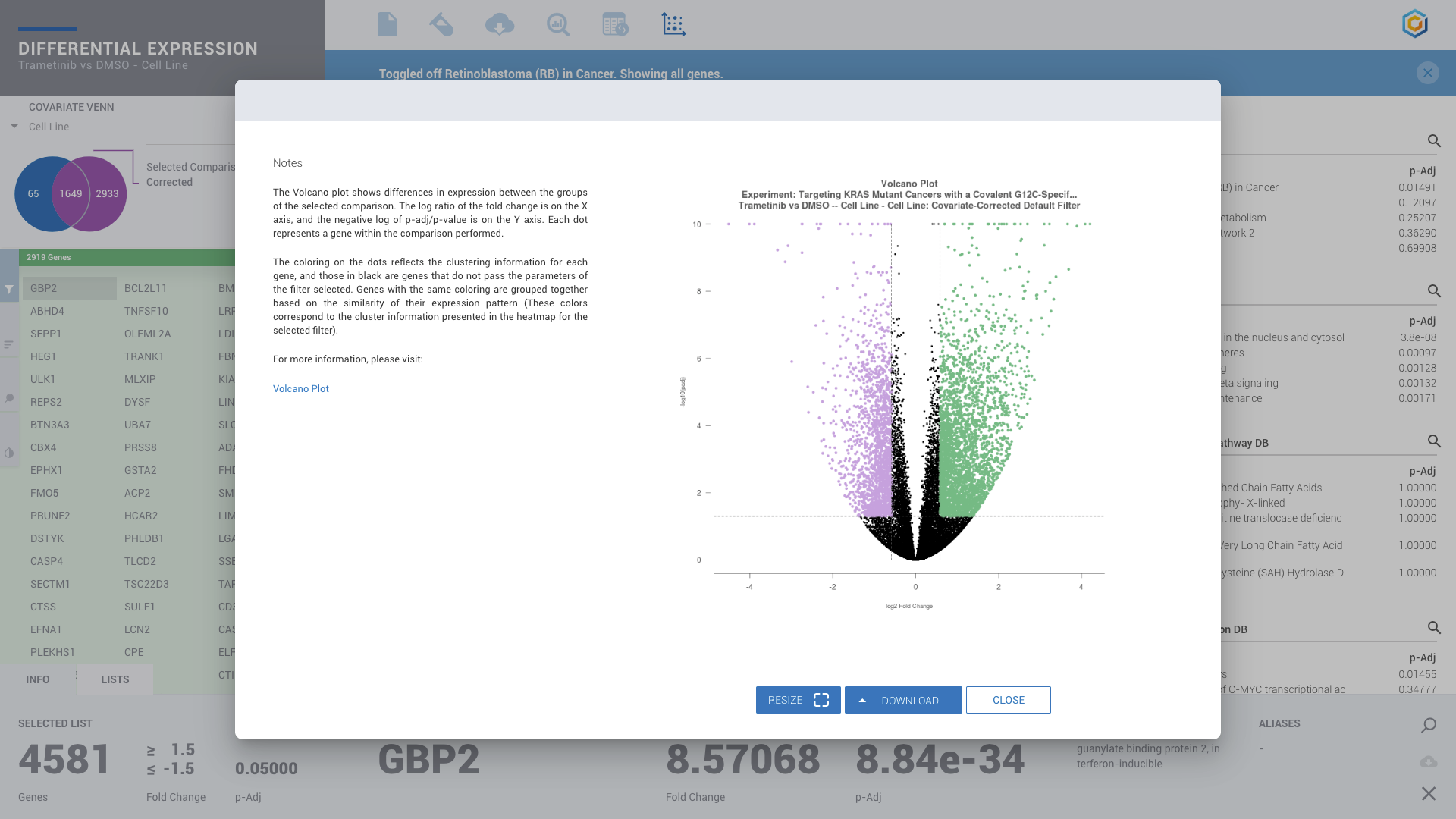
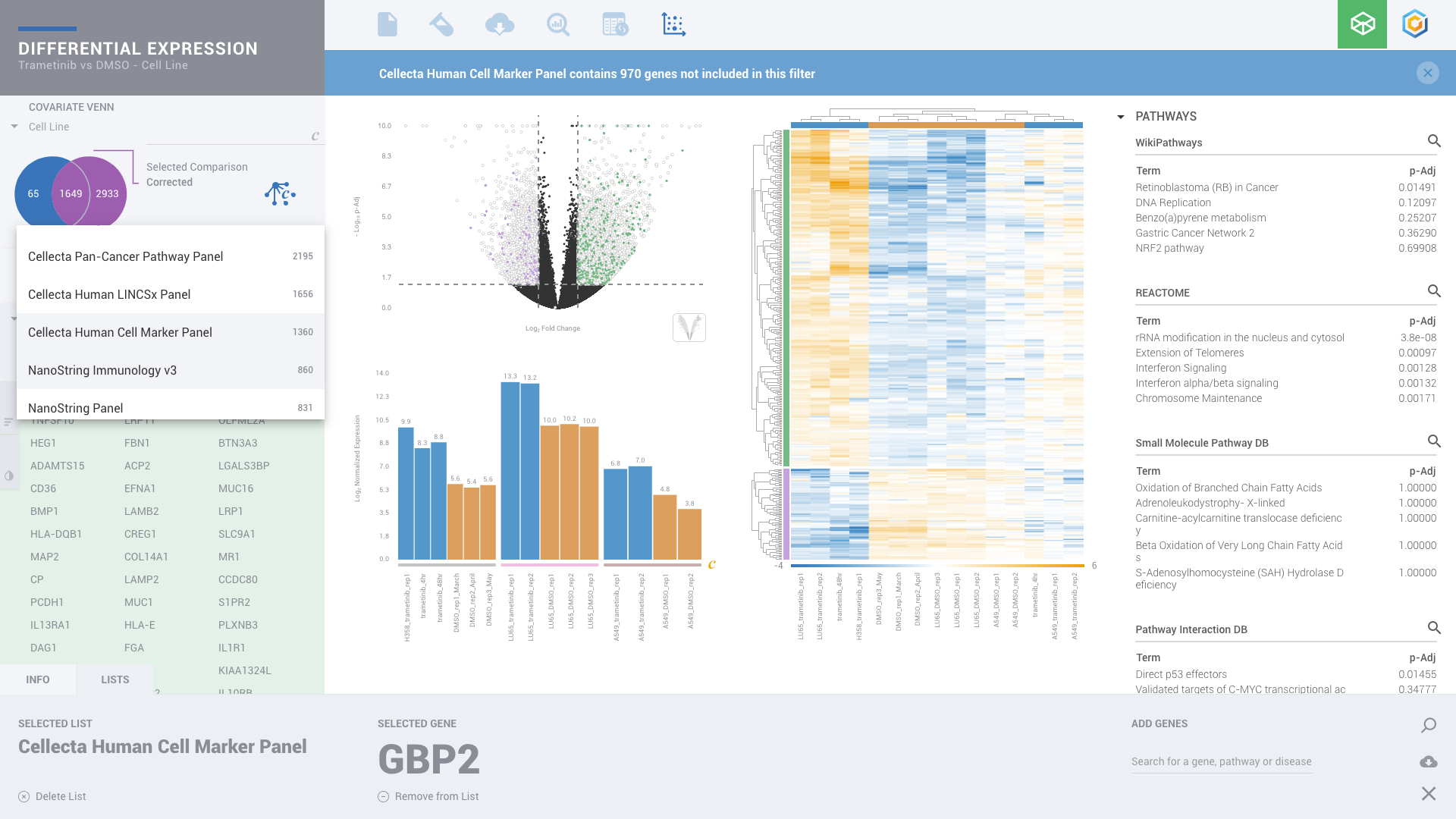
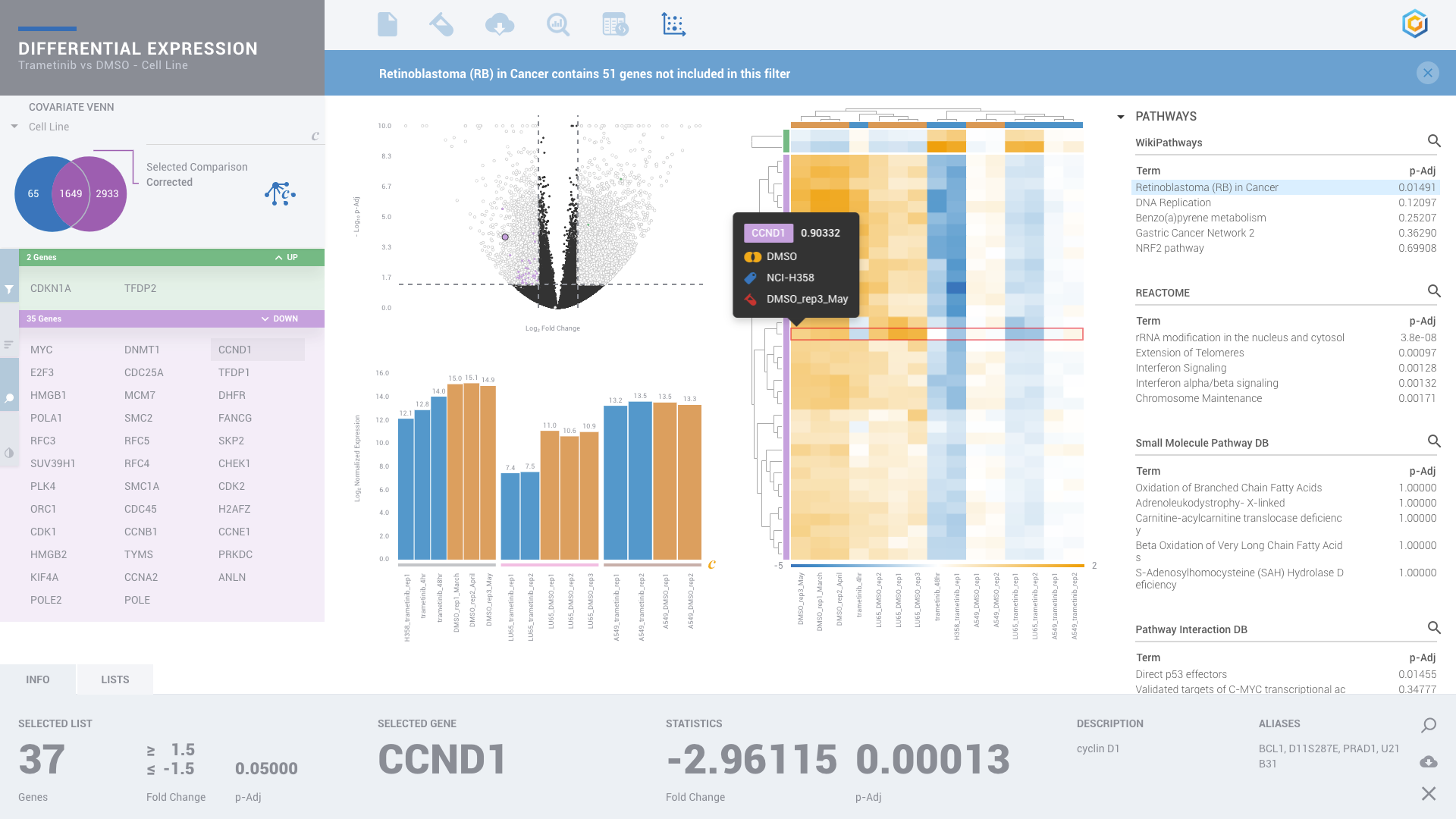
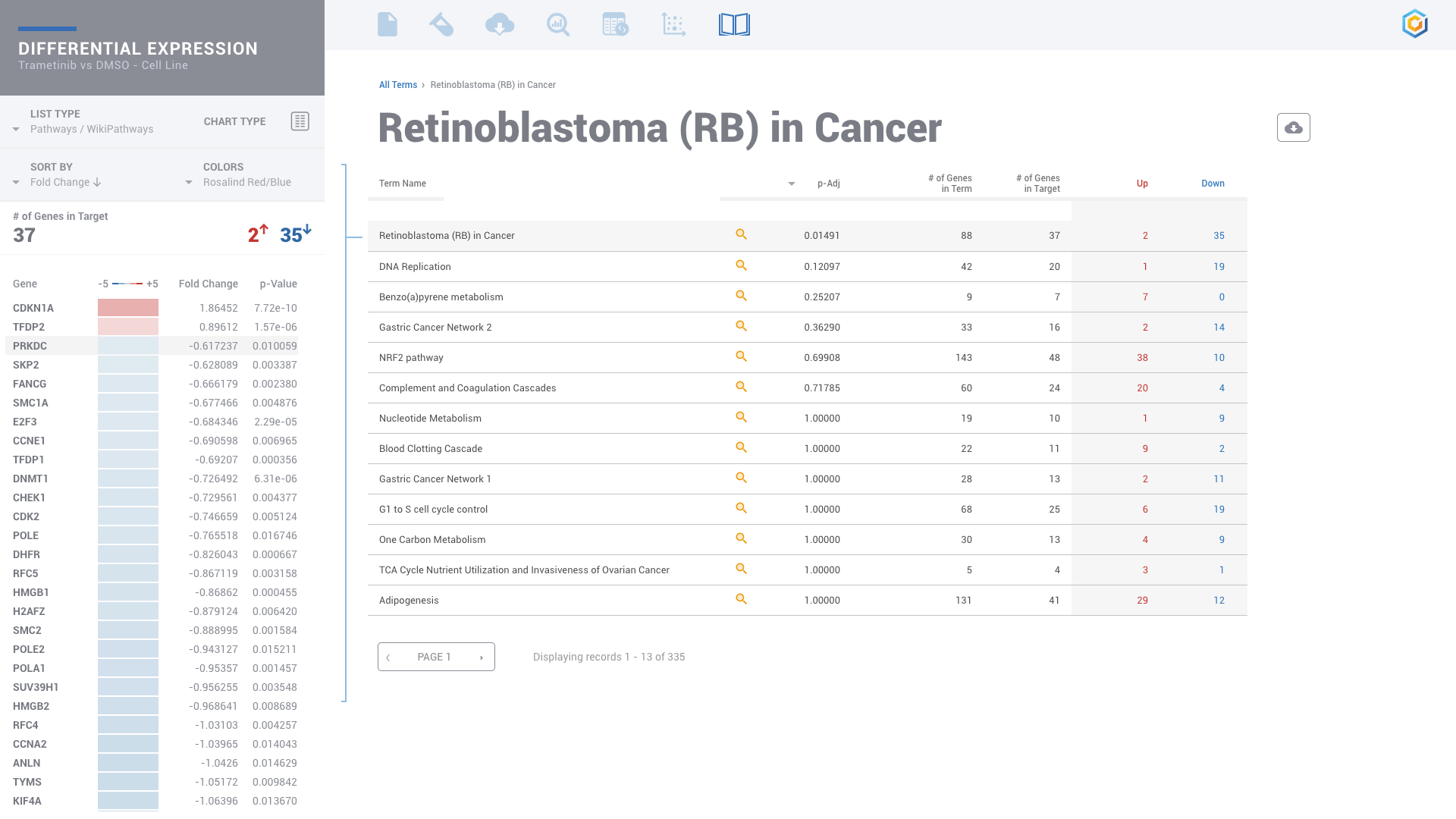
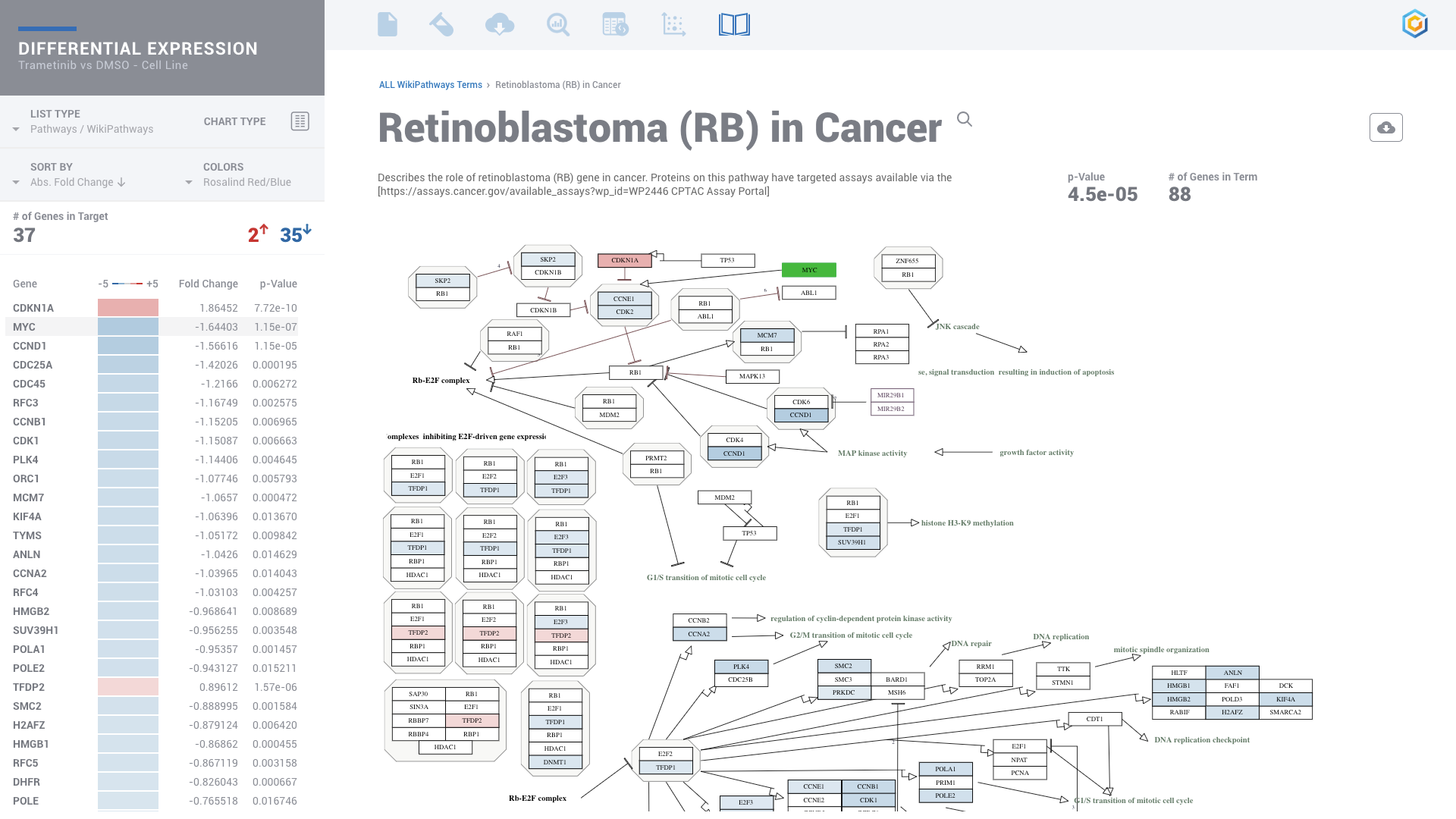
ROSALIND moves beyond the CSV file by providing a comprehensive dashboard for differential expression analysis and interpretation of RNA-seq data. Researchers begin with a list of significant Differentially expressed genes determined by a calculated cut-off filter. Default settings for the filter begin with a fold change of 1.5 upregulated and 1.5 down regulated with a p-Adjust of 0.05. Further adjustments to achieve a significant set of genes are performed by ROSALIND, if needed. Researchers may also create an unlimited set of their own customized filters using fold changes and P value parameters. Convenient on-screen controls are easily accessible for modifying filters, adding covariant corrections, applying gene lists and signatures, and adjusting plot color palettes. The ROSALIND gene expression discovery experience features deep interpretation of top pathways, gene ontology diseases and drug interactions, as rich interactive plots that fill the screen and respond to interactions from the scientist, showing customizable heatmaps, volcano and MA plots as well as box and bar plots.
New comparisons and meta-analysis may be added at any time. Comparisons are created using BioProject attributes. Meta-analyses created can be cross experiments and multi-omic. Each of these perspectives are available within minutes of setup, reducing internal bioinformatic workload and enabling scientists to react fluidly by focusing directly on the science of the experiment.
4. COLLABORATING & SHARING RESULTS
The discovery process rarely ends with a single point of view from a single researcher opinion. ROSALIND Spaces enables true scientist-to-scientist collaboration through virtual data rooms where scientists and collaborators can come together on related datasets anywhere in the world to interactively explore shared experiments much like working with Google Docs. Researchers access a consistent version of the data, without the need to transfer unwieldy files or reinterpret origin files. All changes are interactive, instantly available, and viewable everywhere in the world (as authorized by the organization) with real-time activity feeds and historical reports. Spaces participants can add experiments, explore pathways, change cut-offs, add meta-analyses and add new comparisons all within the shared collaborative environment.
Spaces are virtual meeting rooms where scientists meet with niche experts, clients and supporting teams to maximize the discovery value of every experiment and prepare for the next one.
COMPREHENSIVE GENE EXPRESSION ANALYSIS
HIGHLIGHTS
DESIGNED FOR SCIENTISTS
POWERFUL
EASE OF USE
RICH DATA VISUALIZATION
PATHWAY INTERPRETATION
START FROM FASTQ or PROCESSED DATA
TRUSTED PIPELINES BUILT-IN
SECURITY AND ENCRYPTION


SUPPORTED INSTRUMENTS





SUPPORTED KITS *
FREQUENTLY ASKED QUESTIONS
I am not a bioinformatician. Can I really perform my own analysis?
Absolutely and other scientists just like you run their own analyses on ROSALIND every day. To learn more how to get started, check out the ROSALIND Quick Start Guide here.
What types of Gene Expression experiments are supported?
The ROSALIND Gene Expression discovery experience supports RNA-seq, NanoString gene and protein panels, and Micro-Array (via counts).
What types of input files are supported?
For Gene Expression experiments, FASTQ files and count files are supported. Compressed FASTQs will have faster upload times. Supported file types: .FASTQ, .FASTQ.GZ, .CSV, .TXT, .RCC (NanoString only)
What is an Analysis Unit and how is it used on ROSALIND?
Samples that are processed on ROSALIND require an Analysis Unit to unlock the ROSALIND discovery experience. Analysis Units are already included in most subscriptions on ROSALIND. Additional Analysis Units may be purchased in packs of 10 or 50 from the ROSALIND Store. Analysis Units do not expire. A current subscription is required to utilize Analysis Units. Enterprise Subscriptions provide additional flexibility for high-volume environments. Please contact sales to learn more sales@onramp.bio .
What is considered a Sample?
Any sample that is prepared for processing on an instrument is considered a Sample for ROSALIND. If a Scientist takes two (2) aliquots of an original sample to have replicates and prepares a library for each, this would be considered two (2) Samples on ROSALIND. On the other hand, a Sample may have multiple files associated with it, depending on how sequencing is performed. A single sample may be single-end, paired-end, and also multi-lane and will still be considered as one (1) Sample.
Can I download my results and plots?
Yes. All plots, diagrams, source and results files are downloadable on ROSALIND. Look for the Download buttons to access publication-ready figures as well as to download all experiment datasets.
Do you have an API for programmatic interfacing?
Yes. We provide API integration for Enterprise customers. This allows production teams to automate the upload, processing and distribution of genomic datasets. API integration also includes Single-Sign-On (SSO) support.

Reach Deeper, Explore and Discover More.
Start Your Free Trial
Receive the free ROSALIND Quick Start Guide with your trial