Which Knowledge Bases are included in ROSALIND Pathway Interpretation?
ROSALIND offers best-in-class pathway interpretation including enrichment with over 50 leading knowledge bases
Here's a short list:
- Pathways: WikiPathways, BioPlanet, Reactome, Panther, BIOCYC,
Pathway Interaction Database, and the Small Molecule Pathway Database - MSigDB Pathway Collection: Hallmark, Chemical and Genetic Perturbations, Reactome, BioCarta, Protein Interaction Database
- Oncology & Immunology: Cancer Gene Neighborhoods, Cancer Modules, Oncogenic Signatures, Immunological Signatures
- Transcription & Regulation: MSigDB - Transcription Factor Targets, JASPAR - Transcription Factor Targets, TRRUST - Transcriptional Regulatory Networks, miRNA Targets, miRNA Target Interactions, miRNA Target Predictions, and Chromosome Location
- Diseases: ClinVar, PheWeb, GWAS, DisGeNet, Jensen Diseases
- Drugs: FDA Approved Drugs, Kinase Inhibitors, BROAD Connectivity Map, Computational Drug Signatures, Guide to Pharmacology, Drug-Gene Interaction Database, Drug Matrix - Toxicogenomics Gene Signatures
- Virology: P-Hipster - Pathogen-Host Interactome, GEO - Virus Perturbations
- Ontology Collection: Human Phenotype Ontology, Biological Processes, Molecular Functions and Cellular, Molecular Functions and Cellular Component, GO Molecular Function, GO Biological Processes, and GO Cellular Component, including advanced pruning to eliminate largely redundant terms
- Cell Types & Tissues: Cell Atlas
- Protein Collection: Protein-Protein Interactions, Interpro, Pfam, SMART, GENE3D, Prosite
You can access our Pathway Interpretation by selecting the magnifying glass from any of our recommended pathways on the far right-hand side of your differential expression.
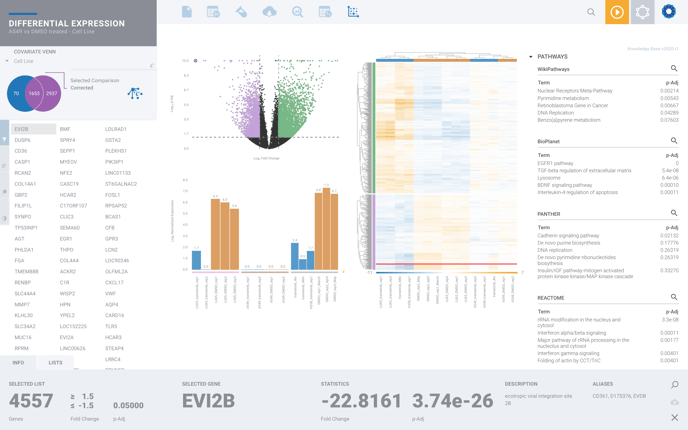
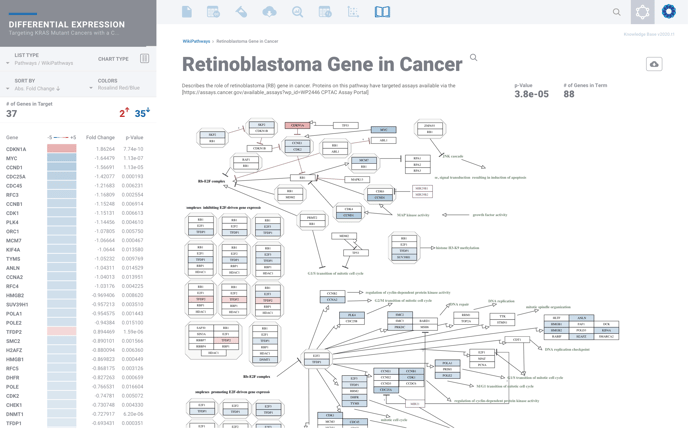
From here you can explore deeper by selecting the golden magnifying glass to view the complete pathway, where available.
/Rosalind_Logo_Primary_RGB.png?width=100&height=75&name=Rosalind_Logo_Primary_RGB.png)