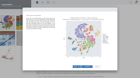
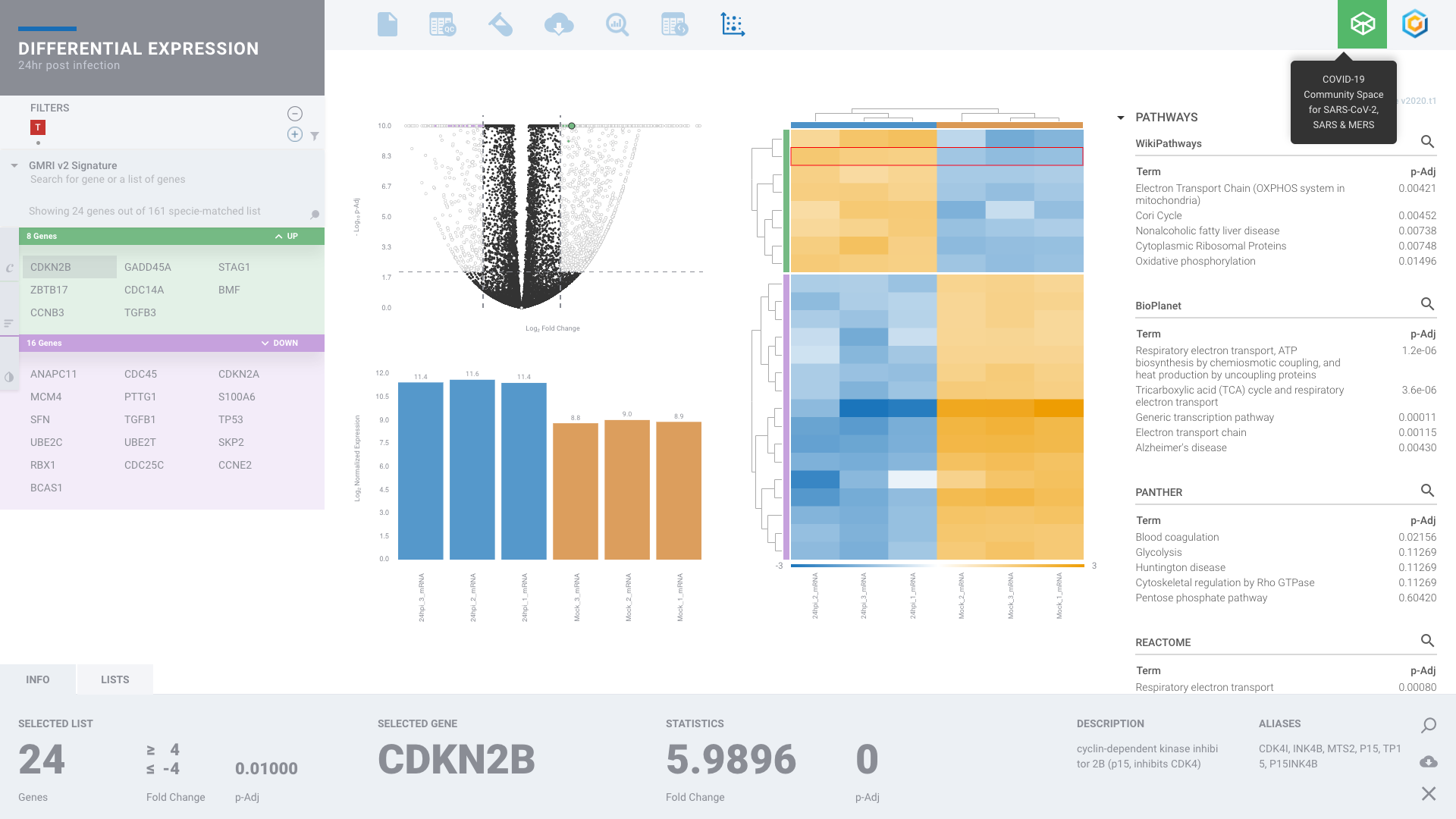
Bulk RNA-seq Gene Expression
DIY Bioinformatics for this genomics staple
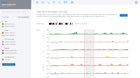
NanoString GeoMx Spatial Transcriptomics
The recommended and preferred solution for best-in-class GeoMx data analysis
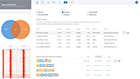
NanoString nCounter Gene and miRNA Expression
Bruker's recommended and preferred solution for best-in-class nCounter data analysis
Single Cell Gene Expression
From FASTQ to cell clusters and beyond
Gene Regulation & Anti-Sense: Small RNA-seq
Small RNA-seq data analysis designed for the biologist
Histone Mark & Transcription Factor: ChIP-seq
Comprehensive ChIP-seq data analysis

Chromatin Accessibility: ATAC-Seq
Genome-wide chromatin accessibility analysis

Sharing & Collaboration
Accelerate teamwork anywhere in the world

Knowledge Graph and Search
Data organized for semantic queries

COVID-19 Diagnostic Monitoring System
SARS-CoV-2 Viral Mutation Tracking
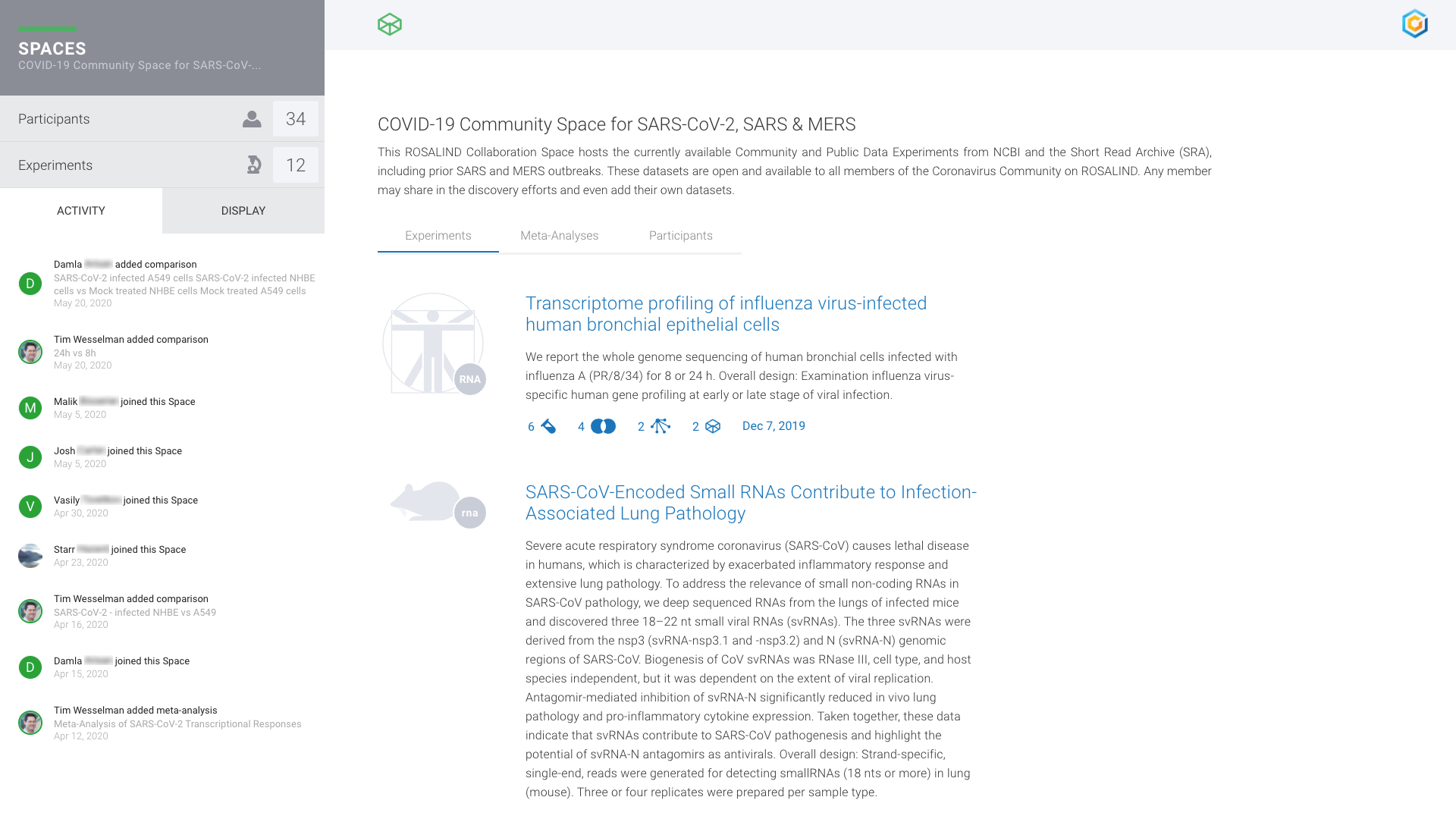
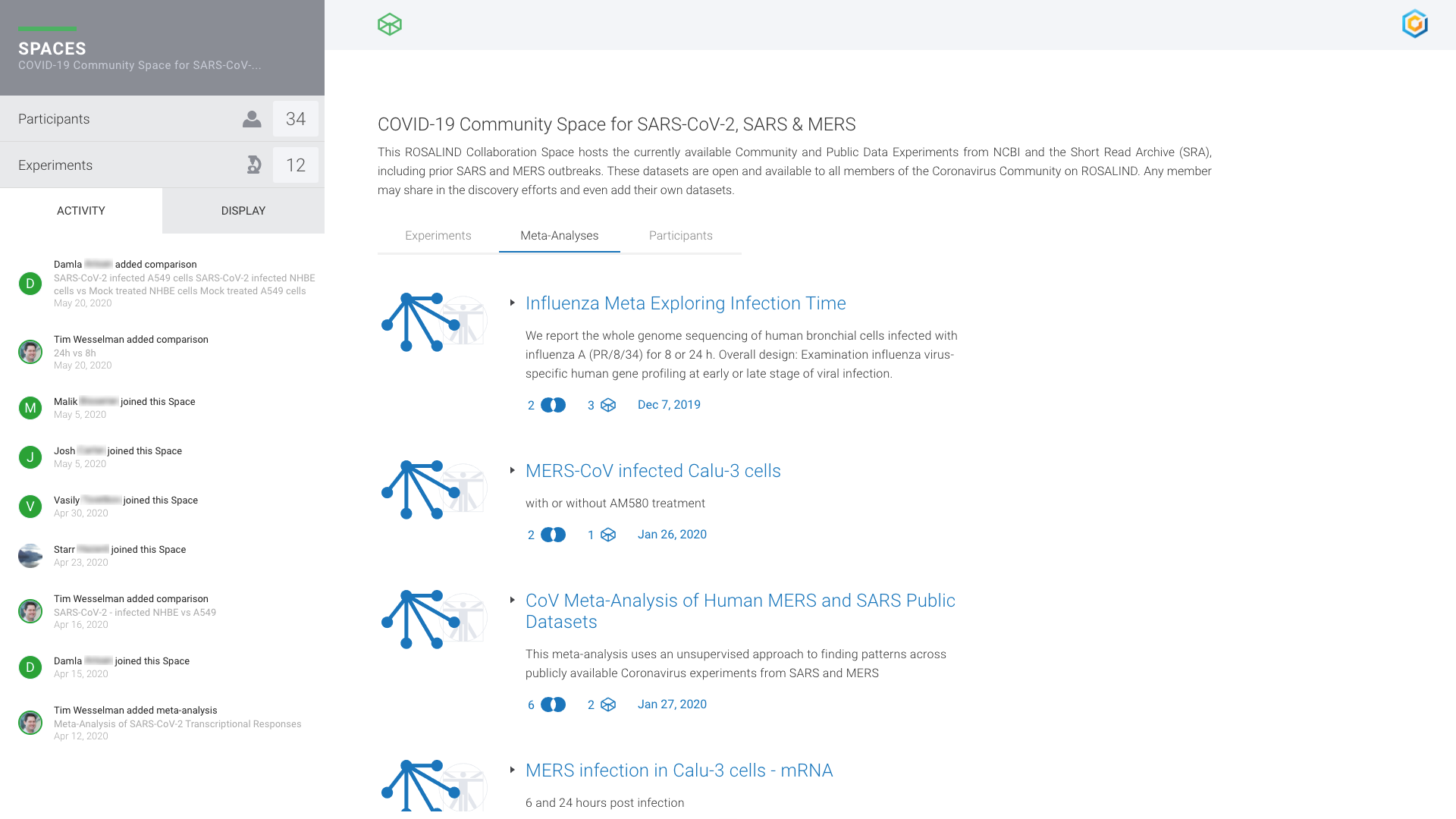
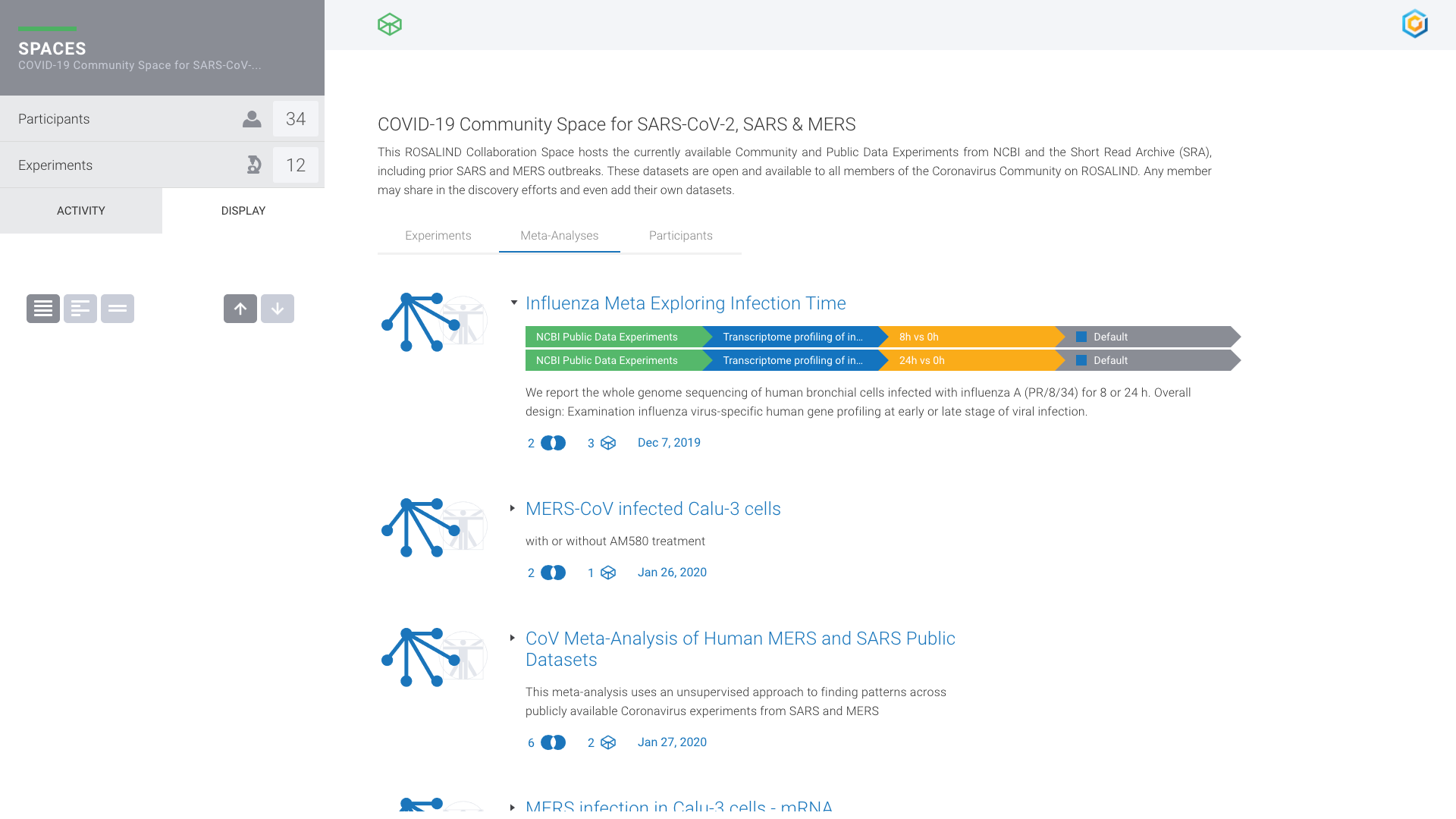
COVID Research Community
Global research analyzing COVID-19 genomic datasets
Immuno Oncology
Making discoveries on IO research
Breast Cancer
Identifying differentially expressed miRNAs in breast cancer
HIV
Transcriptional and genomic profiling study of HIV+ DLBCL
Diabetes
Discover and accelerate T1D
Multi-Omics Dataset
Explore multi-omics datasets in epigenetics research

Single Cell Data
Explore datasets with our Cell Ranger graph-based clustering

nanoString Gene Expression
Analyze your nCounter data in minutes

nanoString Mouse Glial Profiling Panel
Explore neuroinflammatory astrocyte subtypes in the mouse brain

RNA-Seq Data
Get started with bulk RNA-Seq data
Pharma, Biotech, and Academic Researchers
Learn about Enterprise, Professional, and Academic subscriptions
Service Providers
Learn about Service Provider, Contract Research Organization (CRO), and Academic Core Lab licenses