Working with Public Data is important to nearly every Scientist in our community. Unfortunately, it's harder than it should be to work with these valuable datasets. We all see and hear the news each week of exciting discoveries and breakthroughs in leading research, yet accessing and analyzing these Public Datasets, especially from RAW (SRA), requires determination, persistence and more coding skills than should be required of Scientists today.
I'm thrilled to announce that this week, we enabled unlimited NCBI Public Data import and exploration on ROSALIND for our Scientists, Explorers and Directors. This is simply the best way to work with Public Data anywhere and it works like this:
When creating a new experiment, simply choose between Public Data or one of your own datasets.
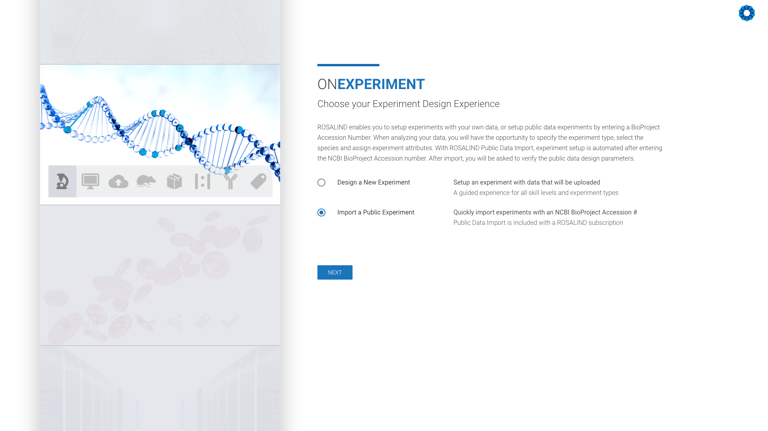
This is included with each of our ROSALIND Plans and does not require the use of any Analysis Units, regardless of the size and number of samples in the experiment.
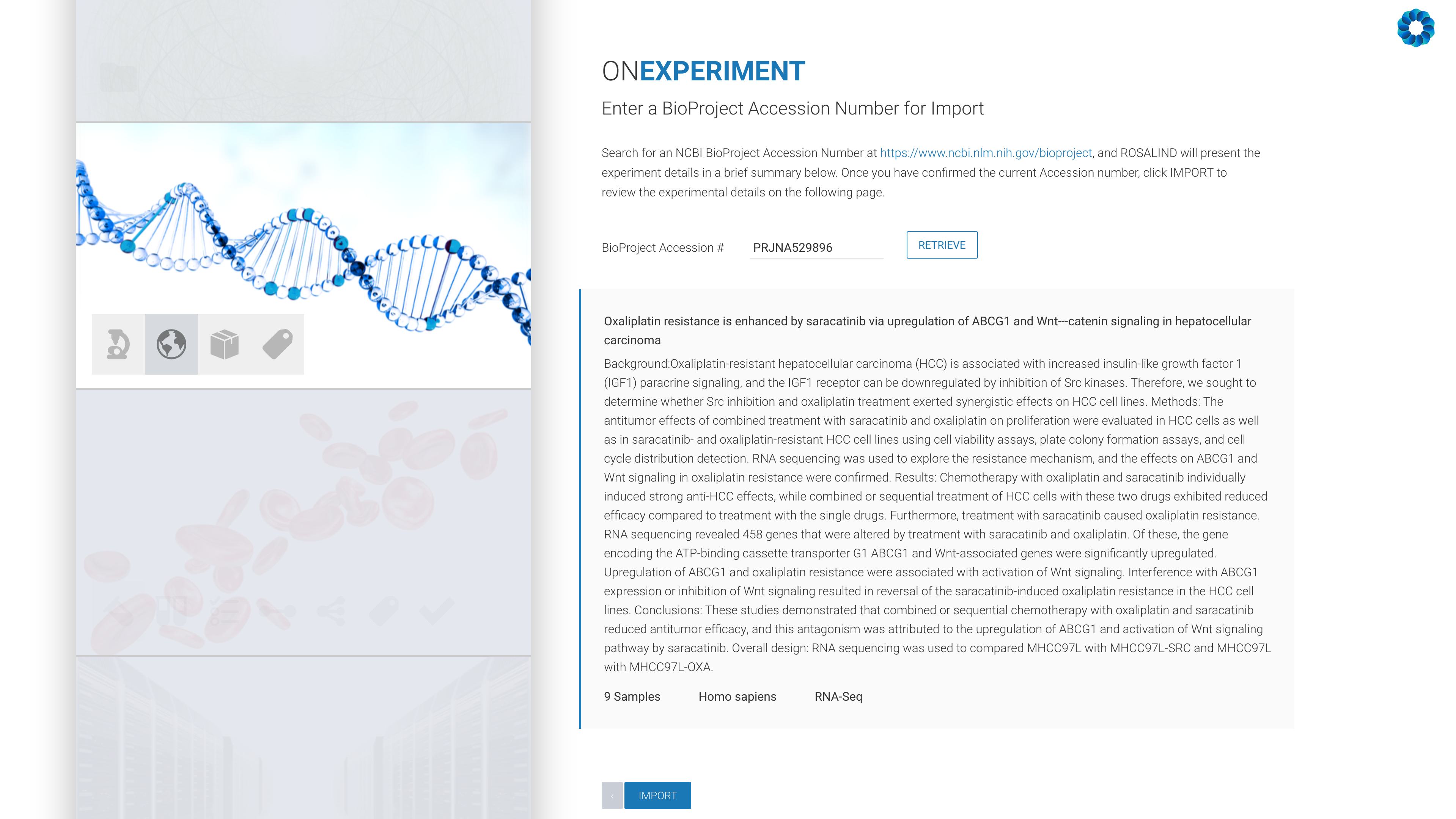
Next, enter an NCBI BioProject Accession number:
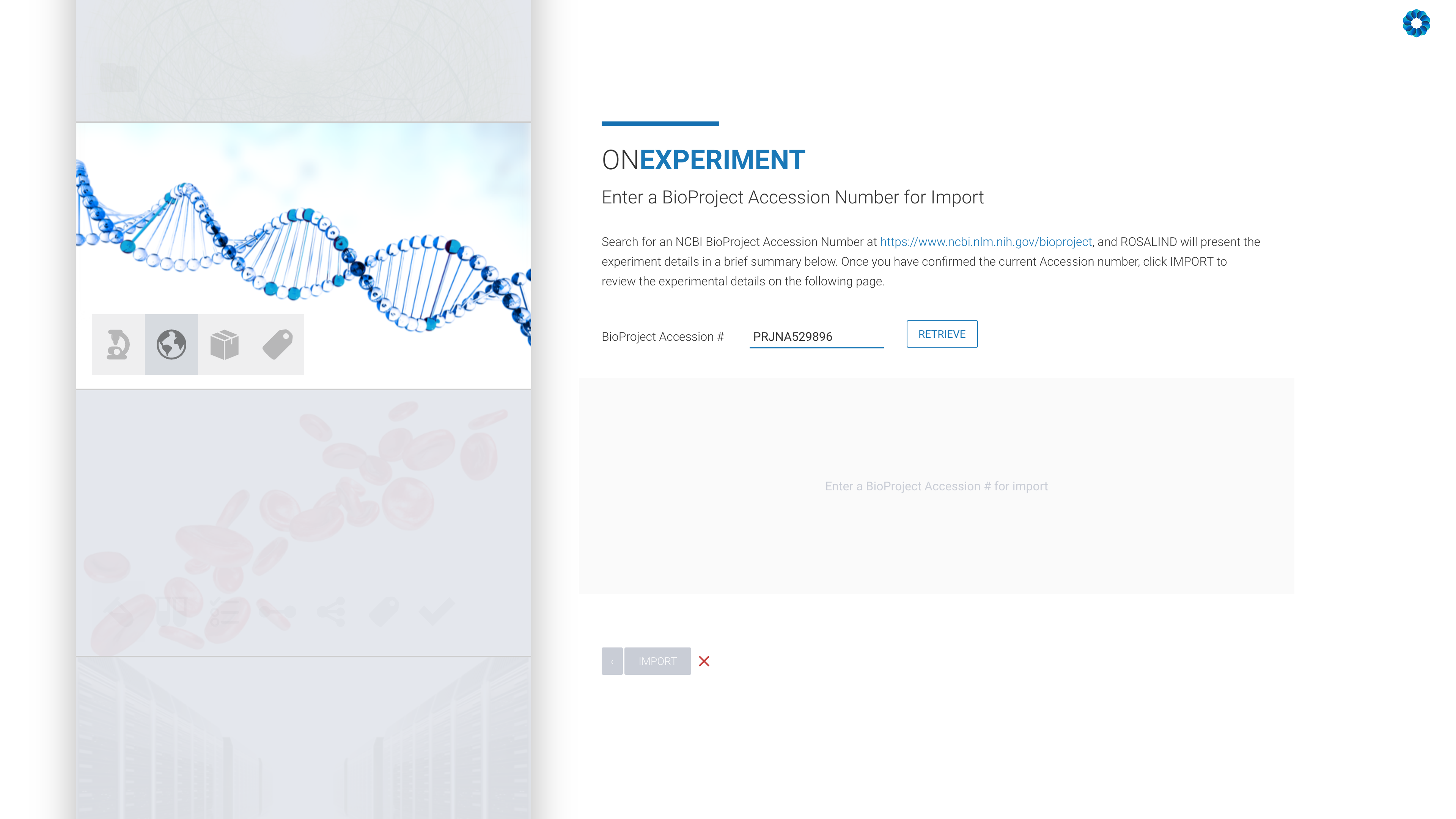
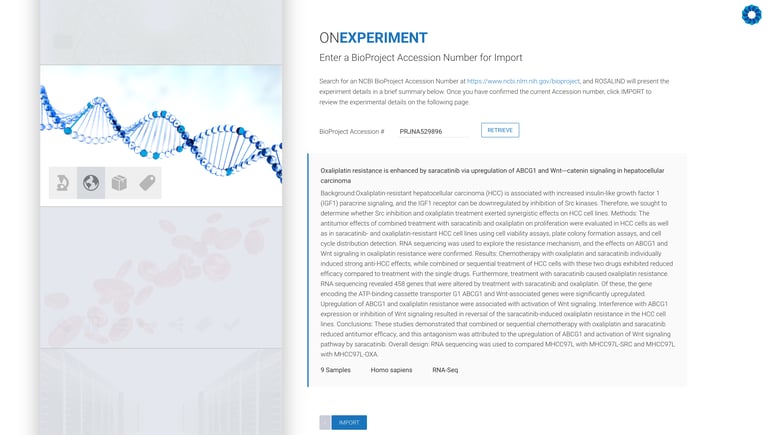
All that remains is the confirmation of the experiment design, including kit selection, setting replicates and defining the desired comparisons (this can also be done after processing has completed).
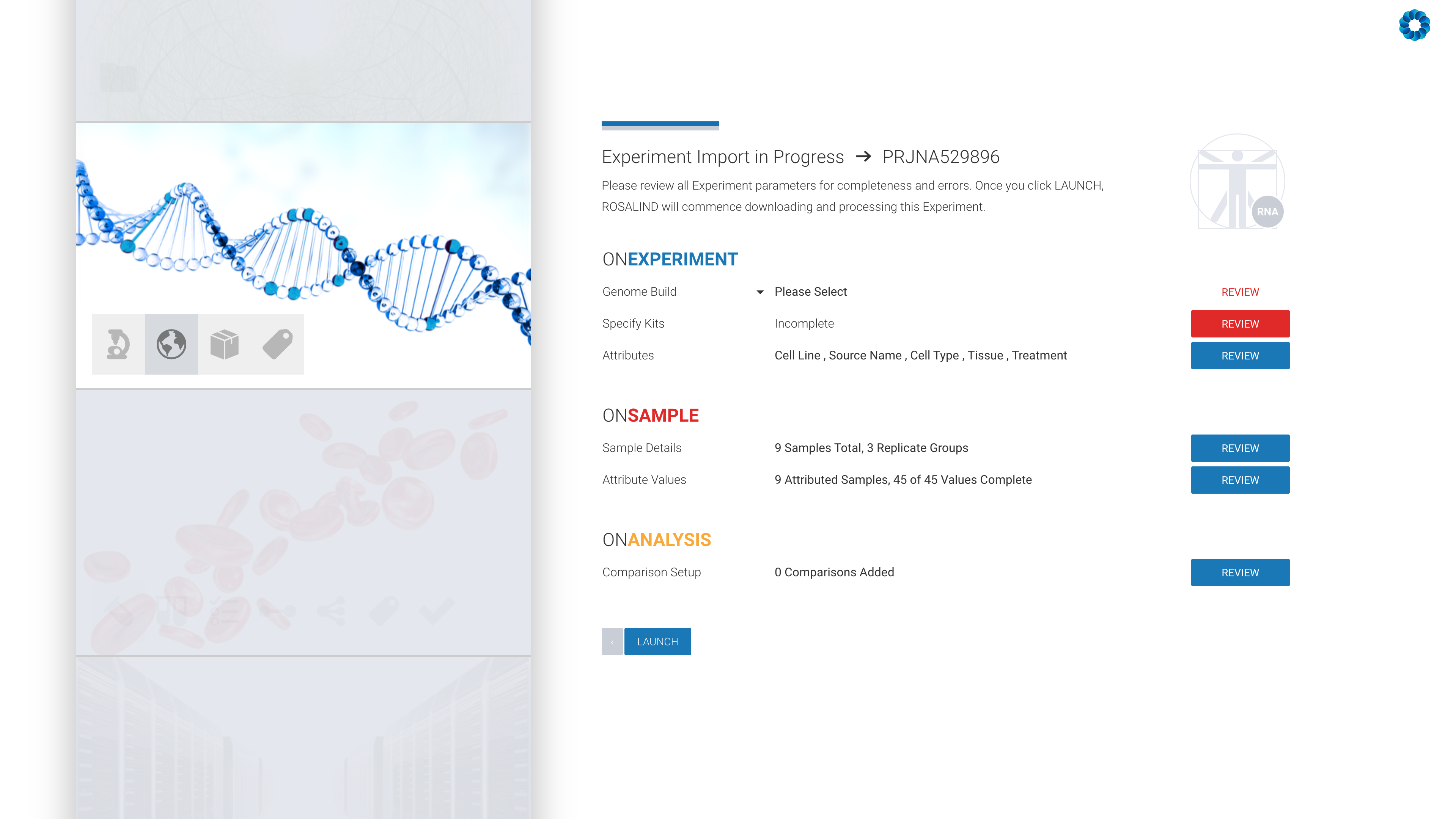
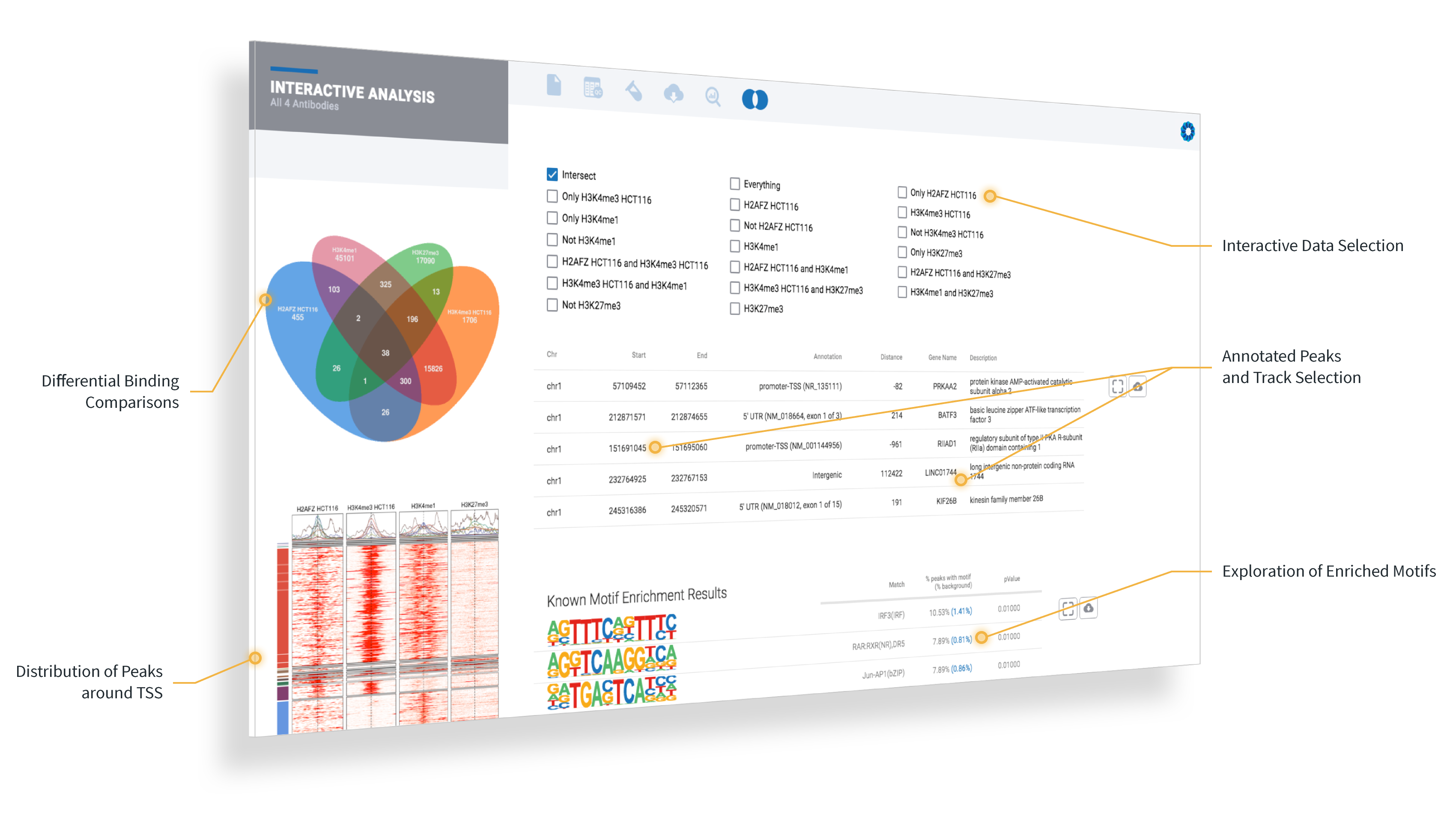
ROSALIND launches the experiment next and completely automates the public data download, data analysis pipelines and prepares beautifully interactive plots and diagrams to explore experiment results with complete pathway enrichment interpretation.
CONCLUSION:
After the analysis completes, the exploration really begins: add new cutoffs or comparisons, explore pathways and gene ontology, create a meta-analysis across comparisons, or to investigate patterns with your own datasets.
I couldn't be more proud of our team and incredible experience ROSALIND now brings to public data exploration. If you don't already have an account or subscription, start a free Scientist trial so you can test this out for yourself.