Welcome to our exciting journey through the world of spatial transcriptomics, an innovative approach transforming the way we conduct research. Spatial analysis, particularly using platforms like GeoMx, opens new avenues for discovering insights that traditional methods may overlook. Today, we dive into one of the most crucial aspects of data analysis: quality control.
You can watch the video, or access the replay below:
Why Quality Control Matters in Spatial Data
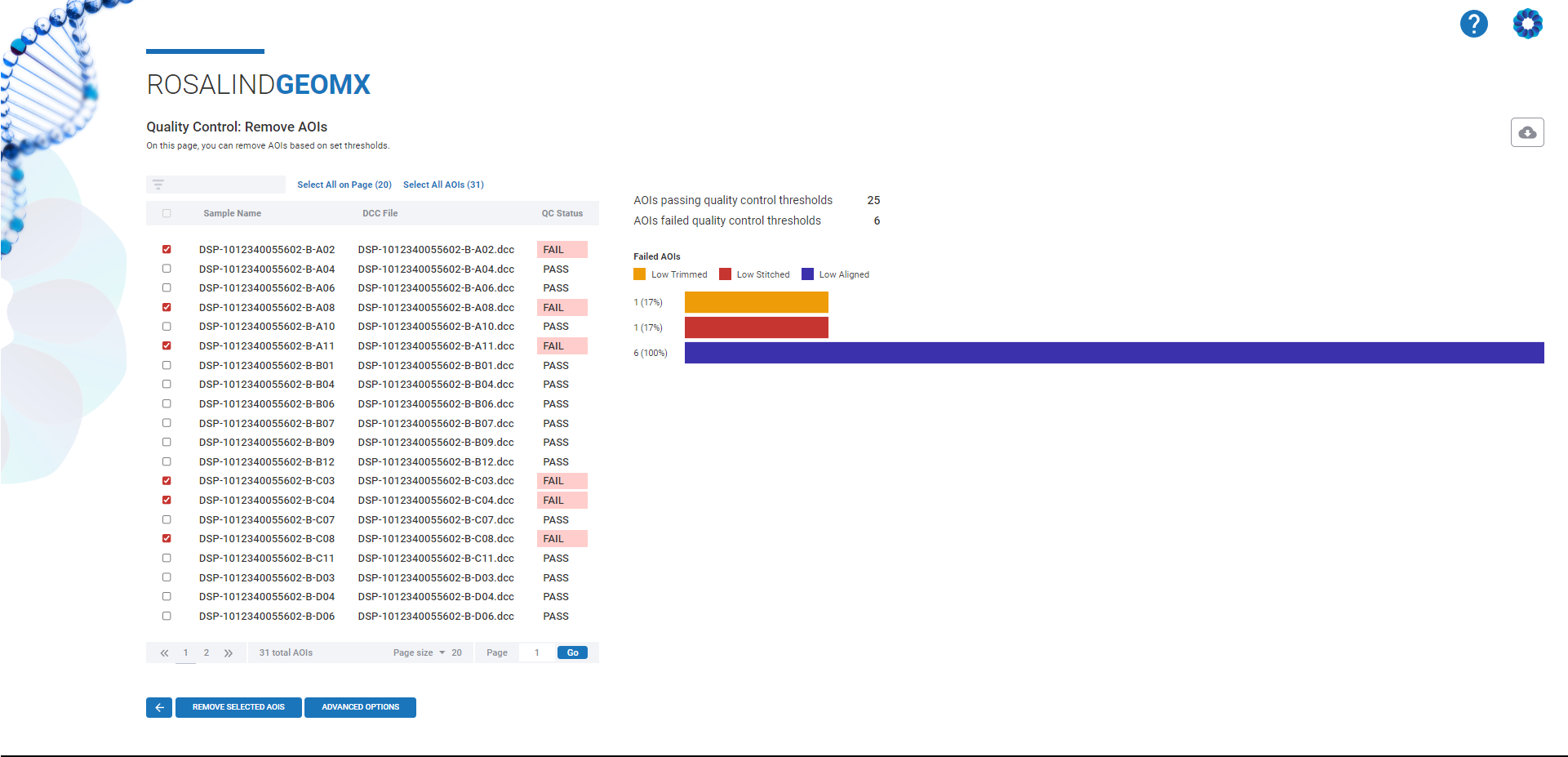
In any research field, ensuring the accuracy and reliability of your data is paramount. Spatial transcriptomics is no exception. Quality control allows researchers to assess the suitability of samples and data points for inclusion in an experiment. For GeoMx WTA data, areas of interest (AOIs) are the primary samples, and identifying “bad” samples early prevents spurious biological interpretations later.
Navigating the Challenges of Spatial Data
Spatial transcriptomics is still a relatively new field, just 5 to 10 years old. While its potential is massive, the technology and software supporting it are constantly evolving. This evolution brings challenges, particularly when it comes to making data analysis user-friendly. Many current tools, such as R and Python, offer flexible and transparent pathways but require advanced expertise, both in coding and spatial statistics.
This is where Rosalind steps in. By creating a more intuitive, transparent, and flexible platform, Rosalind makes spatial data analysis more accessible, even for those without deep coding knowledge.
Breaking Down the QC Process
Let’s dive into the three major steps Rosalind recommends for conducting quality control on GeoMx WTA data:
-
Assess Overall Experiment Quality
The first step is to evaluate the non-template control (NTC) to check for contamination. Contamination at this stage can seriously affect the accuracy of your entire dataset, leading to incorrect interpretations of biological significance. -
Evaluate Sequencing Performance
Once contamination is ruled out, the next step is assessing the sequencing performance. This includes trimming adapter sequences, stitching together matched pairs of data, and aligning reads to known genes. Each step provides a glimpse into whether your sequencing was successful or if issues such as low reads or sequencing errors may have compromised your data. -
Model Dependencies
Finally, look at model-dependent metrics like sequencing saturation and negative probe count. These metrics help ensure that you’ve sequenced your data deeply enough to capture all the diversity within your sample, from low to high expressors.
The Power of Rosalind in QC
Rosalind’s platform not only makes it easier to perform these quality control steps but also provides valuable insights into where issues may arise in your data. For example, Rosalind’s platform helps flag potential contamination, sequence errors, and more, allowing researchers to address these issues before moving forward.
Rosalind’s intuitive design allows researchers to adjust thresholds, view detailed QC parameters, and remove problematic AOIs from datasets—all without needing to dive deep into complex coding platforms. Whether you’re a seasoned expert or new to spatial analysis, Roslin brings transparency and ease to the complex process of data analysis.
Looking Ahead
Our next sessions will continue to explore the intricacies of spatial data analysis, diving into topics like AOI and gene removal, normalization, and spatial differential expression. Stay tuned as we continue to unlock the full potential of spatial transcriptomics and help you make the most of your research with Rosalind.
About Dr. Jessica Noll
Jessica Noll received her Ph.D. from UC Riverside in Biomedical Science. She has expertise in spatial biology, immunohistochemistry, and neuropathology with multiple publications in spatial biology and data analysis. She has previous experience as a NanoString Field Application Scientist, and has a keen understanding and passion for spatial project design and data troubleshooting.












.png)