You can watch the video, or access the replay below:
Spatial biology is transforming the way researchers study complex tissue environments, enabling new insights into disease mechanisms. However, a crucial yet often overlooked part of the data analysis pipeline is normalization—adjusting raw data to ensure accurate comparisons and downstream biological interpretations. In this post, we explore custom normalization methods for GeoMx Whole Transcriptome Analysis (WTA) and discuss how the Rosalind platform simplifies this.
Why is Normalization Essential?
Normalization ensures that technical variations, such as differences in sequencing depth or tissue collection quality, do not obscure the biological signals of interest. If not corrected, these technical biases can lead to misleading results. For example, RNA integrity or probe performance may differ between samples or even within the same tissue, introducing variability unrelated to the biological model being studied.
Normalization Pitfalls
While normalization is essential, it must be done correctly. Overcorrecting data can introduce unnecessary bias, while under-correcting can leave technical noise unaddressed. For instance, some researchers mistakenly apply multiple normalization factors, such as scaling by both surface area and gene expression, which can overfit the data and diminish meaningful biological differences.
Selecting the right normalization strategy requires balancing technical precision and biological relevance—especially when working with technologies like GeoMx, which generates massive datasets through probe hybridization.
The Rosalind platform offers users the flexibility to choose between several normalization methods. Here’s how the most common methods perform:
1. Q3 Normalization-
Uses the third quartile of gene expression to align data across regions of interest (ROIs).
-
Ineffective at accounting for differences in signal-to-noise ratio, resulting in biased downstream interpretations.
-
Adjusts for sequencing depth and RNA composition using the median of ratios method.
-
Moderately effective but still shows bias in some cases, making it less suitable for spatial datasets.
3. Quantile Normalization
-
Aligns distributions of gene expression values across samples by ranking genes and averaging ranks.
-
Corrects for both technical noise and systematic biases, producing more reliable results.
A study published by van Hijfte et al. (2023) analyzed glioma samples using GeoMx WTA. They found that standard Q3 normalization failed to adjust for background noise, leading to skewed data interpretation. After testing various methods, they determined that quantile normalization provided the most consistent results by eliminating technical bias and preserving biological signal.
Using quantile normalization, researchers uncovered meaningful spatial gene signatures that would have been masked by other methods. This demonstrates how choosing the right normalization strategy can unlock new insights from spatial datasets.
---
Simplifying Normalization with Rosalind
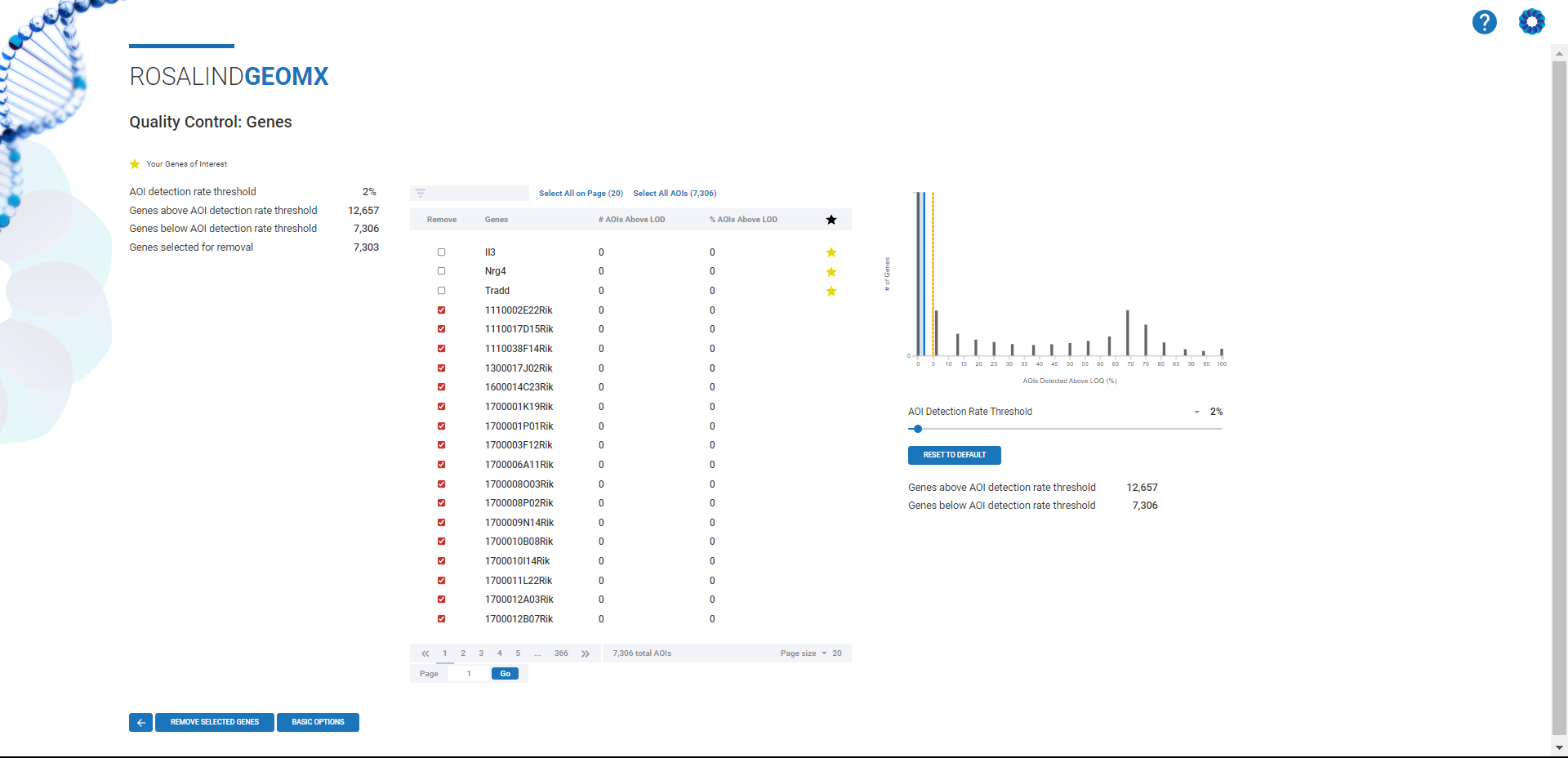
The Rosalind platform takes the complexity out of normalization by providing a non-coding, intuitive interface. Users can select the best-fit normalization method for their datasets through a guided workflow, ensuring consistency across experiments. In addition to offering Q3, DESeq2, and Quantile normalization, Rosalind provides visual tools—such as violin plots with box overlays —to help users assess data quality and make informed decisions.
1. Customization Matters: Different normalization methods are suited to different models and assays.
2. Avoid Overcorrection: Use only the necessary adjustments to preserve true biological differences.
3. Use Visual Tools: Leverage data visualization to validate your normalization approach.
4. Keep Learning: As spatial biology evolves, so will best practices for normalization. Stay informed and adapt your workflows accordingly.
---
Conclusion
Normalization is more than just a technical step in data analysis—it’s a critical process that can shape the outcomes of your research. With platforms like Rosalind, scientists no longer need advanced coding skills to confidently select and apply the right normalization methods. By offering flexibility, transparency, and user-friendly tools, Rosalind helps researchers achieve more accurate and meaningful results in spatial biology.
Interested in learning more? Join our upcoming masterclass on October 31st, where we’ll dive into spatial differential expression and show how Rosalind simplifies the process even further.
About Dr. Jessica Noll
Jessica Noll received her Ph.D. from UC Riverside in Biomedical Science. She has expertise in spatial biology, immunohistochemistry, and neuropathology with multiple publications in spatial biology and data analysis. She has previous experience as a NanoString Field Application Scientist, and has a keen understanding and passion for spatial project design and data troubleshooting.