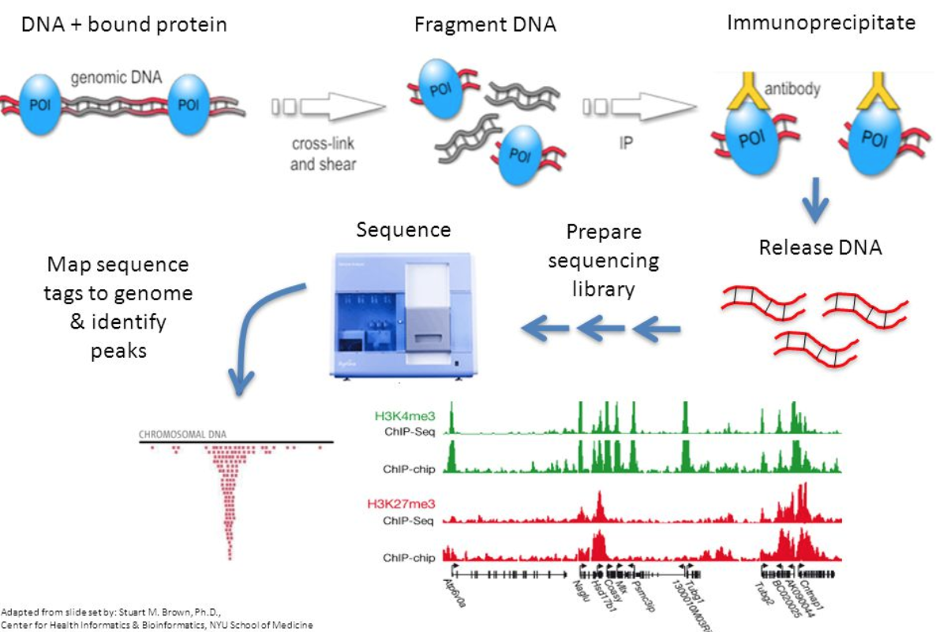
FIVE STEPS TO SUCCESS WITH CHIP-SEQ DATA ANALYSIS
ROSALIND simplifies data analysis and works like a data hub interconnecting every stage of data interpretation. The ROSALIND DNA-Protein interactions discovery experience enables visual exploration and self-investigation of experiment results to give researchers the freedom to explore peak overlaps and differential binding. The peak overlap experience provides visibility into the presence of DNA-Protein interactions regions anywhere in the genome and across multiple comparison groups. The differential binding experience focuses on DNA-Protein interactions on the proximal promoter region of genes regions within the transcription start site (TSS) and gene promoter regions. Based on the differential binding in their promoter region, each gene is displayed with visualization similar to gene expression, including heatmaps, volcano plots, MA plots, bar graphs and box plots. Within this interactive environment, one may adjust cut-offs, add comparisons and even find patterns across multiple datasets and experiment types - such as ChIP-seq, ATAC-seq or RNA-seq multi-omics analyses. There are five easy steps to performing ChIP-seq data analysis on ROSALIND.
1. EXPERIMENT DESIGN
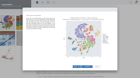
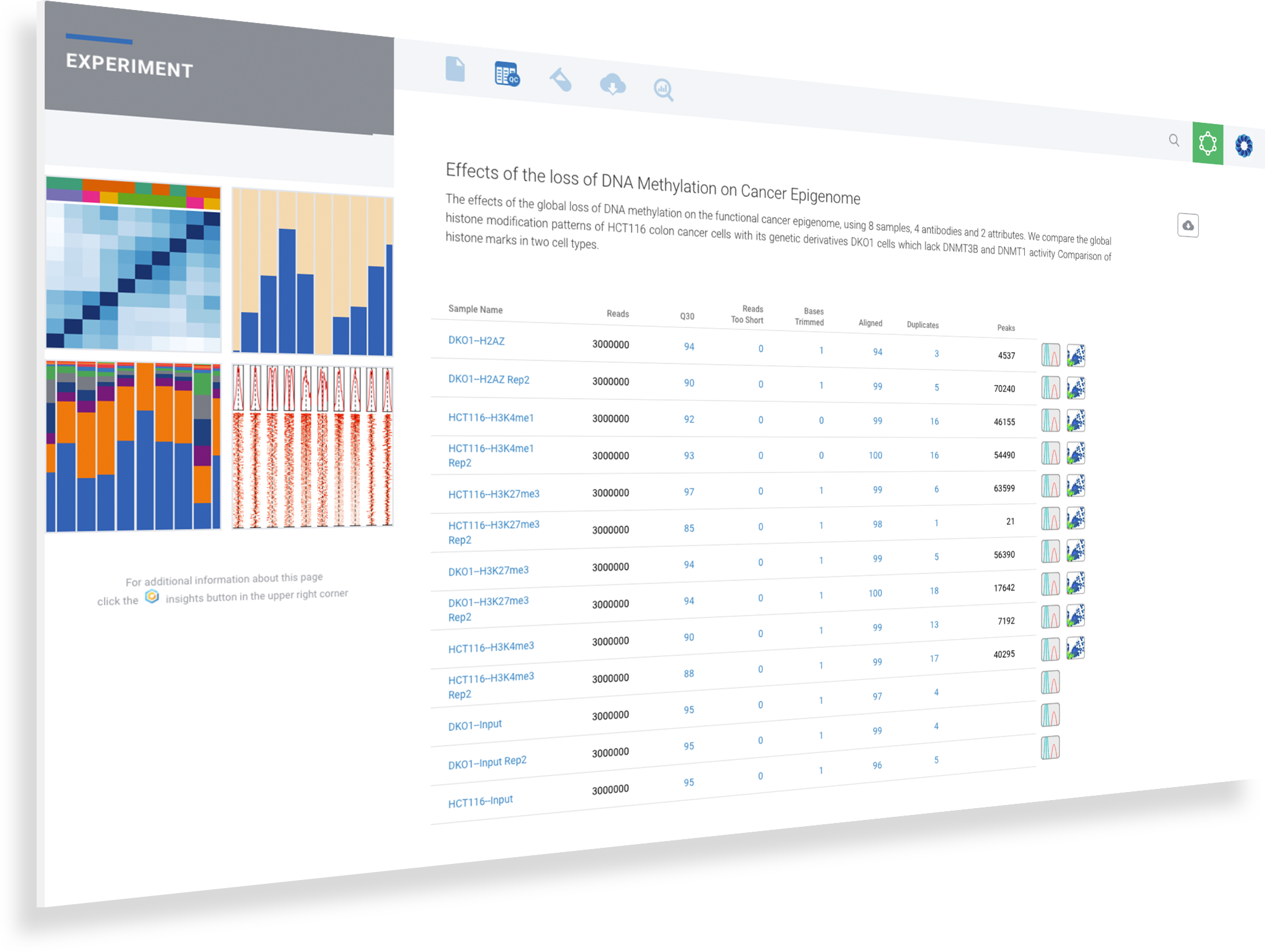
Starting a ChIP-seq data analysis begins with creating a new experiment and capturing the experiment design. ROSALIND walks through the key aspects of an experiment in a guided experience to record biological objectives, sample attributes and analysis parameters. These details become the basis of the experiment discovery dashboard. Researchers who publish papers and work with NCBI public data know the importance of natively supporting NCBI data models. ROSALIND fully supports the NCBI BioProject and BioSample models for metadata assignment and sample attribute descriptions. ROSALIND also enables scientists to create custom attributes to describe biological behaviors in terms relevant to the experiment. The setup of comparisons is simplified by describing and annotating samples using these familiar terms. This methodology minimizes the risk of differential accessibility errors when selecting samples for comparison.
For ChIP-seq data analysis, ROSALIND analyzes the raw FASTQ files produced by high throughput sequencing. ROSALIND streamlines data analysis using an advanced pipeline for analysis that includes intelligent quality control with automatic contamination detection, identification of binding sites or chromatin modification regions and deep pathway interpretation of the genes close by. Visit the technical specifications section to learn more about the ROSALIND ChIP-seq data analysis pipeline and available reference materials.
For proper ChIP-seq results, an analysis pipeline must adjust for sample preparation and proprietary differences in library preparation kits used in the experiment. Not only is the kit selection important for targeting and capturing the desired binding regions, but the analysis pipeline also adjusts and optimizes for the kit’s unique characteristics, such as presence of strandedness, strand direction, any unique molecular identifiers (UMIs) as well as the adapters used. ROSALIND integrates and supports a broad library of sample and library preparation kits, automatically calibrating each analysis with the appropriate details. ROSALIND will also need to know the list of antibodies against proteins and/or histone marks used in the experiment To learn more about supported kits and antibodies, visit the technical specifications section. Featured kits and instrument partners are also listed below.
2. CHIP-SEQ QUALITY CONTROL
Researchers must be confident in the quality control phase before gathering insights from an ChIP-seq experiment, otherwise, the results of the analysis should not be trusted. Biology’s mysteries are elusive and complex. Time should not be lost chasing corrective measures for outliers, contamination, swapped samples and the many other errors that can occur in the course of a well-designed experiment.
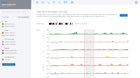
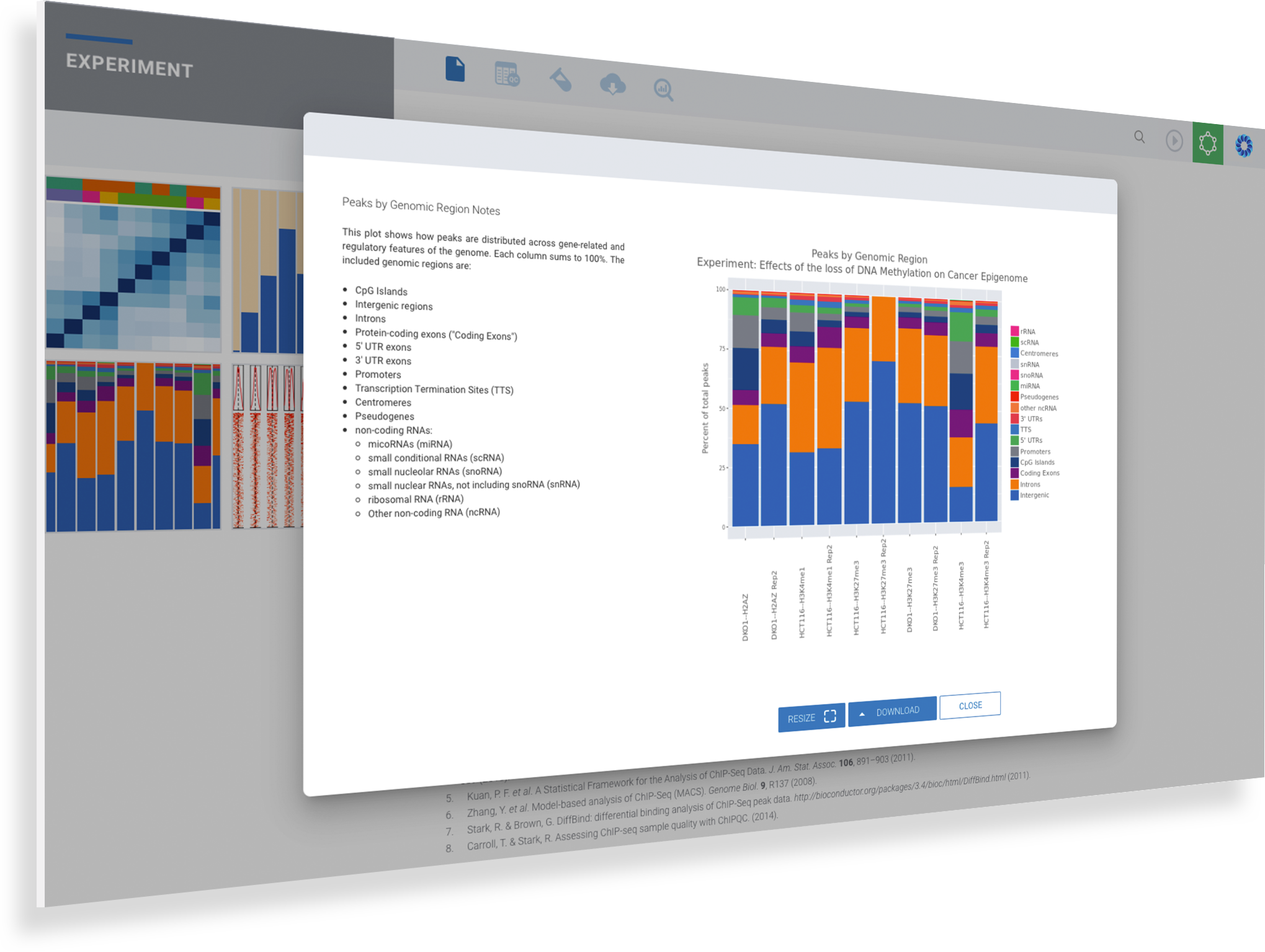
Some of the most important Quality Control metrics to verify are Q30 scores, alignment rates, duplicate rates, number of opened regions detected, sample correlation, fraction of reads in peaks, genomic regions and TSS plot for all samples. When ROSALIND detects low alignment, non-aligning reads are evaluated for possible contamination. For best results with Illumina sequencers, Q30 values should exceed 85% with alignment rates over 80% for the target species. Additional QC metrics, such as duplicate rates, should be less than 25% with fewer than 10% of reads trimmed. Researchers can eliminate offending samples and the deleterious effects on results by identifying the sample as an outlier and move confidently into the discovery and exploration phase of results interpretation.
ROSALIND Quality Control Intelligence identifies potential data quality issues and triages the data before presenting the results. This eliminates the need for researchers to be experts in Sequencing quality control issues. Learn how researchers gain confidence in their results through Quality Control Intelligence.
3. UNLOCKING RESULTS
After a researcher has reviewed the quality control phase the interactive presentation of results is ready to begin. The next step is to unlock the experiment. ROSALIND calculates the quantity of Analysis Units (“AU”) required to unlock the results. This is generally 1 AU per single-sample FASTQ file for ChIP-seq experiments, however, this may differ based on counts files or other experiment parameters. Account balances and quick links for acquiring more AU are directly accessible from the unlock screen. To learn more about Analysis Units, check out the Q&A in the section below, or visit the ROSALIND Store.
4. ANALYSIS & DISCOVERY
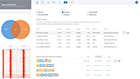
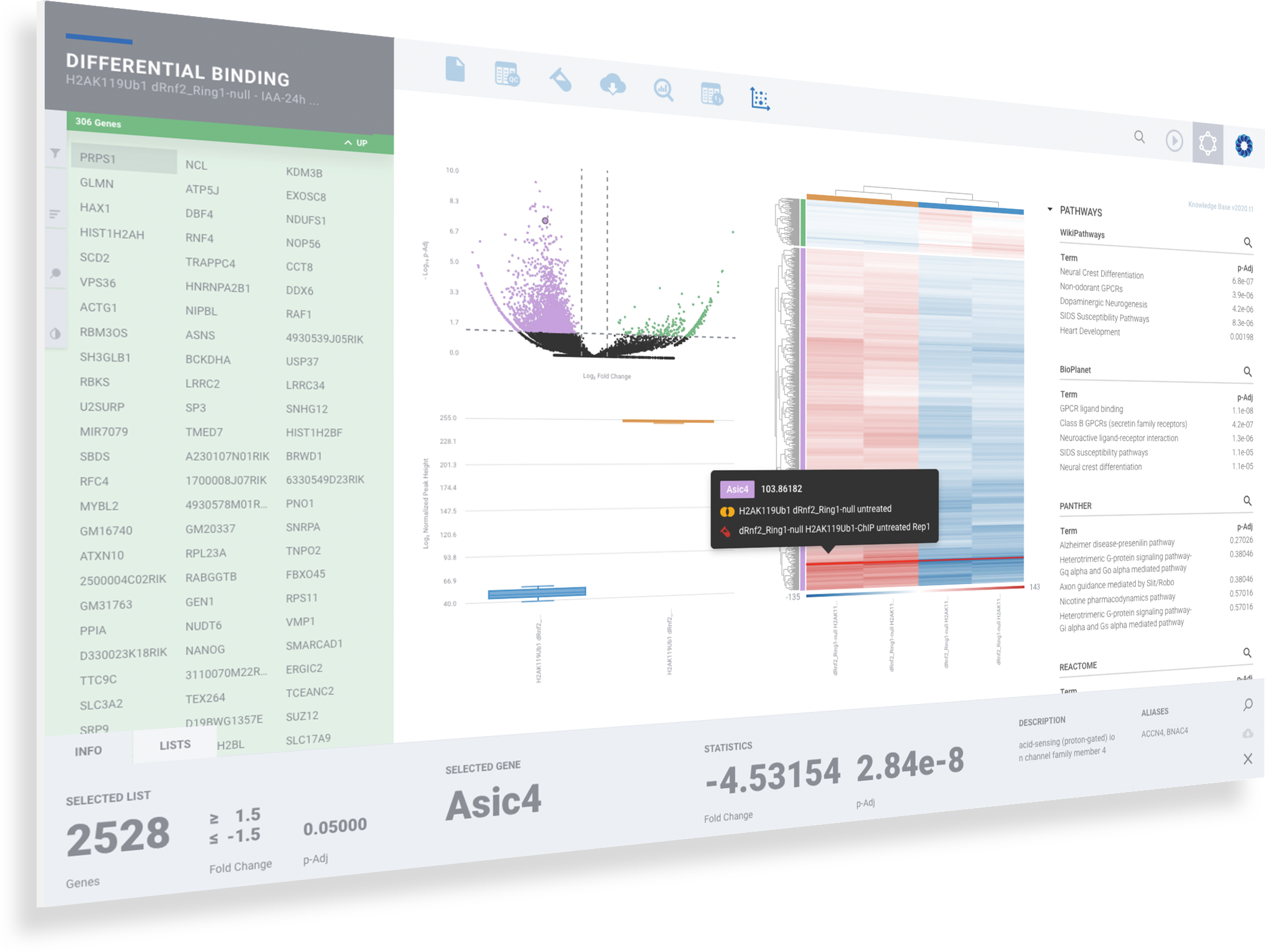
A typical ChIP-seq analysis provides a list of differentially binding regions or histone modifications, generally in the form of a massive and obtuse CSV file. Unfortunately, this often results in more questions than answers for scientists. Multiple applications may also need to be used to generate this CSV file. Such applications often have a wide range of complexity with non-standard input/output formats, many of which are command-line tools requiring advanced knowledge in programming — an exercise well beyond the level of most biologists.
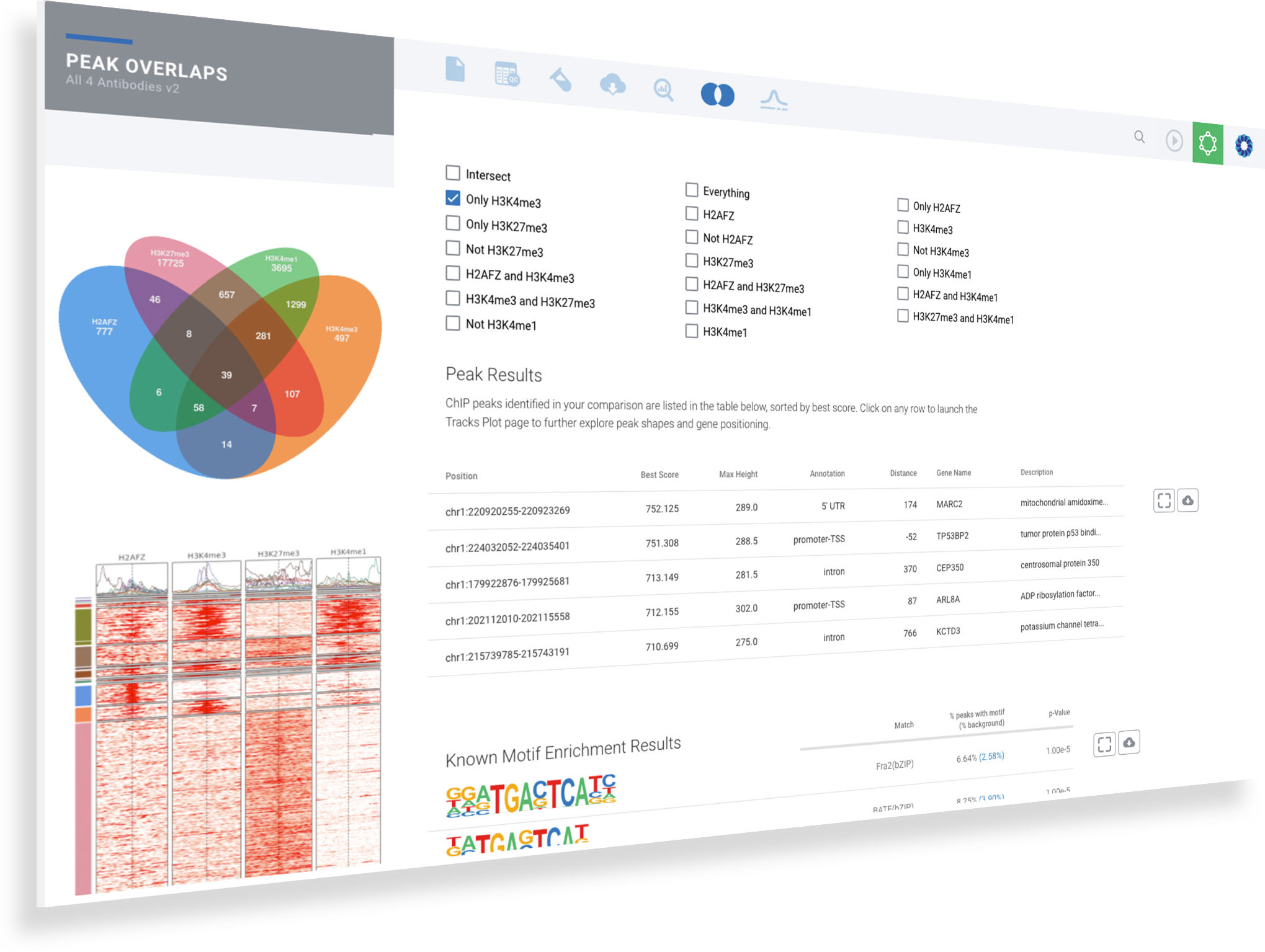
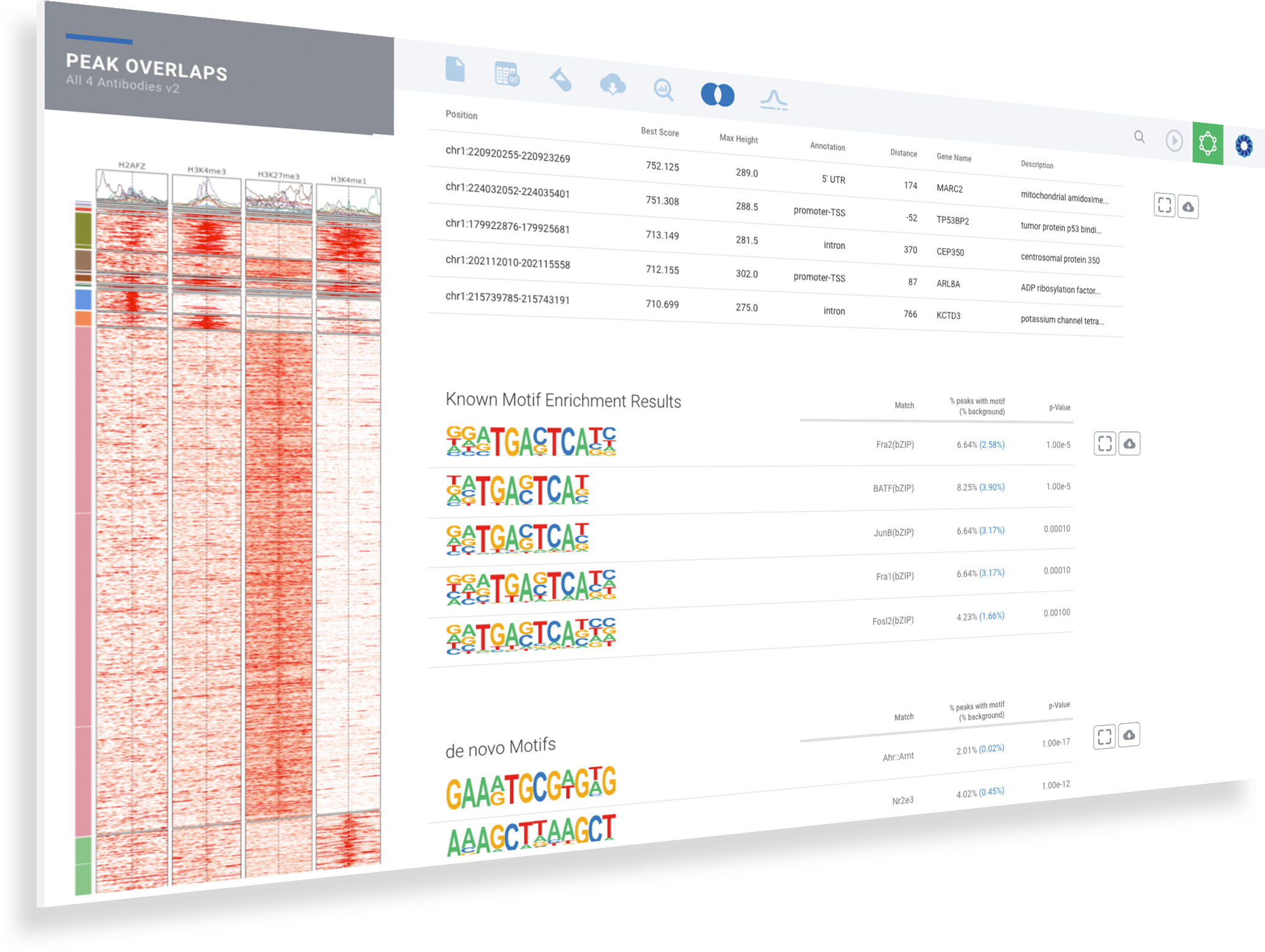
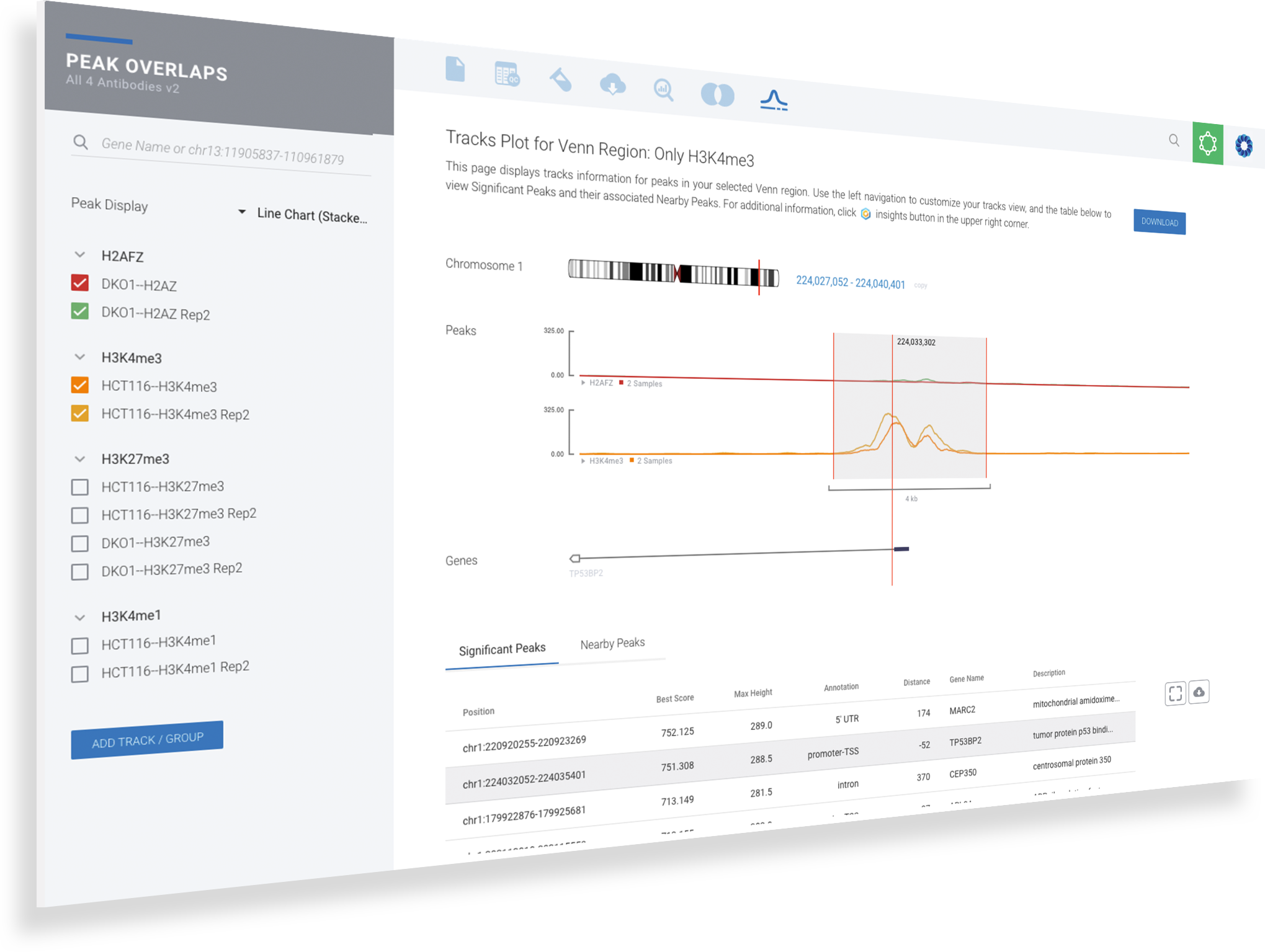
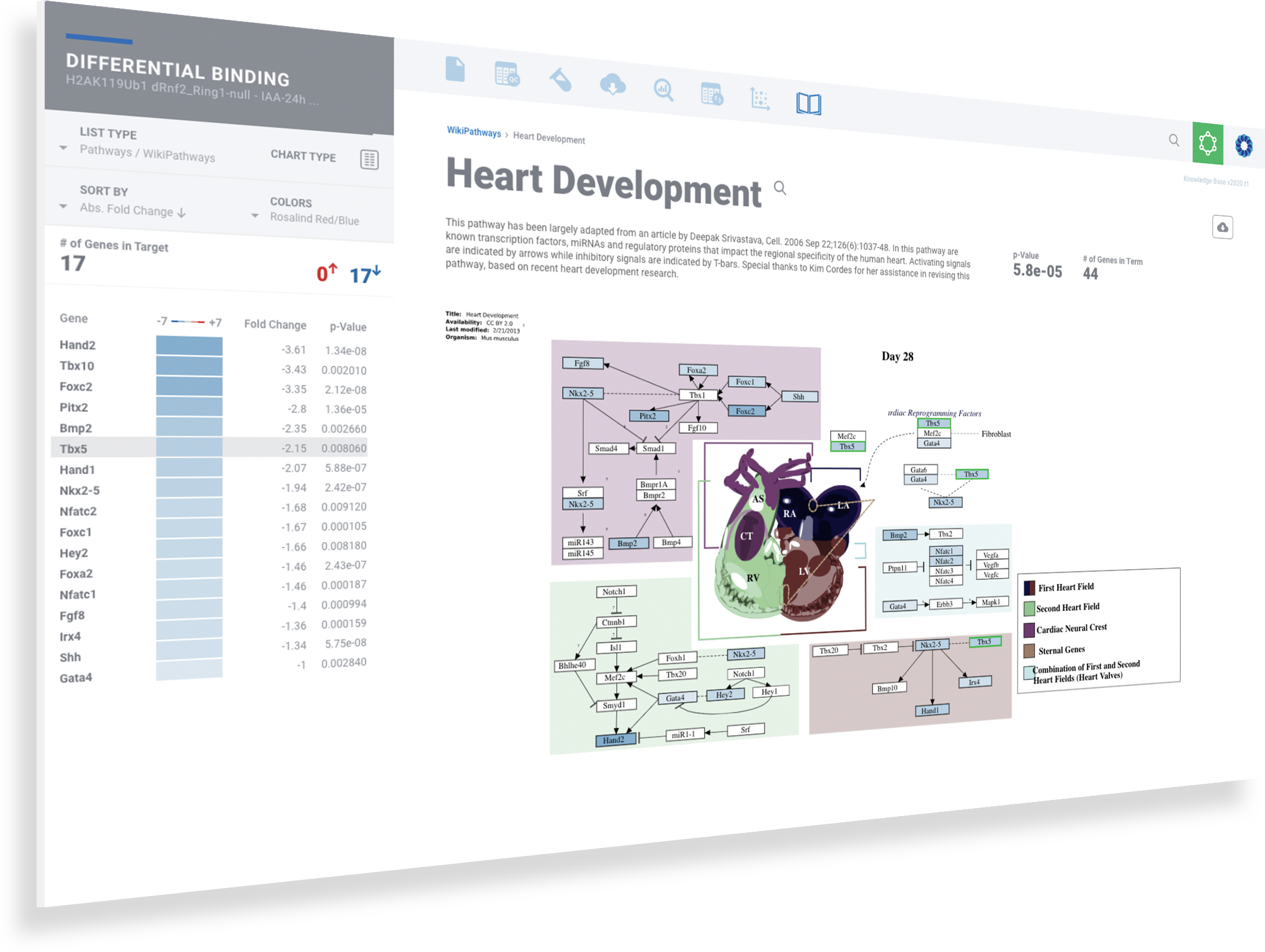
ROSALIND moves beyond the CSV file by providing a comprehensive dashboard for differential binding and interpretation of ChIP-seq data. Researchers begin with a list of significant differentially binding regions determined by a calculated cut-off filter. Default settings for the filter begin with a fold change of +/- 1.5 with a p-Adjust lower than 0.05. Further adjustments to achieve a significant set of regions are performed by ROSALIND if needed. Researchers may also create an unlimited set of their own customized filters using fold changes and P-value parameters. Convenient on-screen controls are easily accessible for modifying filters, applying gene lists and signatures, and adjusting plot color palettes. The ROSALIND differential binding and histone modifications experience features deep interpretation of top pathways, gene ontology diseases, and drug interactions, as rich interactive plots that fill the screen and respond to interactions from the scientist, showing customizable heatmaps, volcano and MA plots as well as box and bar plots.
New comparisons and meta-analysis may be added at any time. Comparisons are created using BioProject attributes. Meta-analyses created can be cross experiments and multi-omic. Each of these perspectives are available within minutes of setup, reducing internal bioinformatic workload and enabling scientists to react fluidly by focusing directly on the science of the experiment.
5. COLLABORATION & DATA SHARING
The discovery process rarely ends with a single point of view from a single researcher’s opinion. ROSALIND Spaces enables true scientist-to-scientist collaboration through virtual data rooms where scientists and collaborators can come together on related datasets anywhere in the world to interactively explore shared experiments much like working with Google Docs. Researchers access a consistent version of the data, without the need to transfer unwieldy files or reinterpret origin files. All changes are interactive, instantly available, and viewable everywhere in the world (as authorized by the organization) with real-time activity feeds and historical reports. Spaces participants can add experiments, explore pathways, change cut-offs, add meta-analyses and add new comparisons all within the shared collaborative environment.
Spaces are virtual meeting rooms where scientists meet with niche experts, clients and supporting teams to maximize the discovery value of every experiment and prepare for the next one.






















/Rosalind_Logo_Primary_RGB.png)